A single amino acid in the reverse transcriptase domain of hepatitis B virus affects virus replication efficiency
- PMID: 11689664
- PMCID: PMC114769
- DOI: 10.1128/JVI.75.23.11827-11833.2001
A single amino acid in the reverse transcriptase domain of hepatitis B virus affects virus replication efficiency
Abstract
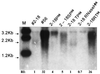
To explore functional domains in the hepatitis B virus (HBV) polymerase, two naturally occurring HBV isolates (56 and 2-18) with 98.7% nucleic acid sequence homology but different replication efficiencies were studied. After transfection into HepG2 cells, HBV DNA isolated from intracellular virus core particles was much higher in 56-transfected cells than in cells transfected with 2-18. The structural basis for the difference in replication efficiency between these two isolates was studied by functional domain gene substitution. The complete polymerase (P) gene and its gene segments coding for the terminal protein (TP), spacer (SP), reverse transcriptase (RT), and RNase H in 2-18 were separately replaced with their counterparts from 56 to construct full-length chimeric genomes. Cell transfection analysis revealed that substitution of the complete P gene of 2-18 with the P gene from 56 slightly enhanced viral replication. The only chimeric genome that regained the high replication efficiency of the original 56 isolate was the one with substitution of the RT gene of 2-18 with that from 56. Within the RT region, amino acid differences between isolates 2-18 and 56 were located at positions 617 (methionine versus leucine), 652 (serine versus proline), and 682 (valine versus leucine). Point mutation identified amino acid 652 as being responsible for the difference in replication efficiency. Homologous modeling studies of the HBV RT domain suggest that the mutation of residue 652 from proline to serine might affect the conformation of HBV RT which interacts with the template-primer, leading to impaired polymerase activity.
Figures




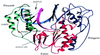
Similar articles
-
A putative new domain target for anti-hepatitis B virus: residues flanking hepatitis B virus reverse transcriptase residue 306 (rtP306).J Med Virol. 2007 Jun;79(6):676-82. doi: 10.1002/jmv.20835. J Med Virol. 2007. PMID: 17457904
-
Mutational analysis revealed that conservation of hepatitis B virus reverse transcriptase residue 306 (rtP306) is crucial for encapsidation of pregenomic RNA.FEBS Lett. 2007 Feb 6;581(3):558-64. doi: 10.1016/j.febslet.2007.01.024. Epub 2007 Jan 18. FEBS Lett. 2007. PMID: 17254572
-
Variable influence of mutational patterns in reverse-transcriptase domain on replication capacity of hepatitis B virus isolates from antiviral-experienced patients.Clin Chim Acta. 2011 Jan 30;412(3-4):305-13. doi: 10.1016/j.cca.2010.10.028. Epub 2010 Nov 5. Clin Chim Acta. 2011. PMID: 21056552
-
Hepatitis B viruses: reverse transcription a different way.Virus Res. 2008 Jun;134(1-2):235-49. doi: 10.1016/j.virusres.2007.12.024. Epub 2008 Mar 12. Virus Res. 2008. PMID: 18339439 Review.
-
Comparisons of the HBV and HIV polymerase, and antiviral resistance mutations.Antivir Ther. 2004 Apr;9(2):149-60. Antivir Ther. 2004. PMID: 15134177 Review.
Cited by
-
Discovery and Selection of Hepatitis B Virus-Derived T Cell Epitopes for Global Immunotherapy Based on Viral Indispensability, Conservation, and HLA-Binding Strength.J Virol. 2020 Mar 17;94(7):e01663-19. doi: 10.1128/JVI.01663-19. Print 2020 Mar 17. J Virol. 2020. PMID: 31852786 Free PMC article.
-
Heat shock protein 90-independent activation of truncated hepadnavirus reverse transcriptase.J Virol. 2003 Apr;77(8):4471-80. doi: 10.1128/jvi.77.8.4471-4480.2003. J Virol. 2003. PMID: 12663754 Free PMC article.
-
Novel evidence suggests Hepatitis B virus surface proteins participate in regulation of HBV genome replication.Virol Sin. 2011 Apr;26(2):131-8. doi: 10.1007/s12250-011-3190-0. Epub 2011 Apr 7. Virol Sin. 2011. PMID: 21468936 Free PMC article.
-
Paradoxical HBsAg and anti-HBs coexistence among Chronic HBV Infections: Causes and Consequences.Int J Biol Sci. 2021 Mar 11;17(4):1125-1137. doi: 10.7150/ijbs.55724. eCollection 2021. Int J Biol Sci. 2021. PMID: 33867835 Free PMC article. Review.
-
In vitro drug susceptibility analysis of hepatitis B virus clinical quasispecies populations.J Clin Microbiol. 2007 Oct;45(10):3335-41. doi: 10.1128/JCM.00272-07. Epub 2007 Aug 8. J Clin Microbiol. 2007. PMID: 17687019 Free PMC article.
References
-
- Ago H, Adachi T, Yoshida A, Yamamoto M, Habuka N, Yatsunami K, Miyano M. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Struct Fold Des. 1999;7:1417–1426. - PubMed
-
- Ayola B, Kanda P, Lanford R E. High level expression and phosphorylation of hepatitis B virus polymerase in insect cells with recombinant baculoviruses. Virology. 1993;194:370–373. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

