An N-terminal domain of herpes simplex virus type Ig E is capable of forming stable complexes with gI
- PMID: 11689673
- PMCID: PMC114778
- DOI: 10.1128/JVI.75.23.11897-11901.2001
An N-terminal domain of herpes simplex virus type Ig E is capable of forming stable complexes with gI
Abstract
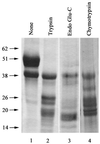
Using limited proteolytic analyses, we show that gE present in soluble herpes simplex virus type 1 gE-gI complexes is cleaved into a C-terminal (CgE) and an N-terminal (NgE) domain. The domain boundary is in the vicinity of residue 188 of mature gE. NgE, but not CgE, forms a stable complex with soluble gI.
Figures



Similar articles
-
UL13 protein kinase of herpes simplex virus 1 complexes with glycoprotein E and mediates the phosphorylation of the viral Fc receptor: glycoproteins E and I.Virology. 1998 Feb 1;241(1):37-48. doi: 10.1006/viro.1997.8963. Virology. 1998. PMID: 9454715
-
The extracellular domain of herpes simplex virus gE is indispensable for efficient cell-to-cell spread: evidence for gE/gI receptors.J Virol. 2005 Sep;79(18):11990-2001. doi: 10.1128/JVI.79.18.11990-12001.2005. J Virol. 2005. PMID: 16140775 Free PMC article.
-
Herpes simplex virus gE/gI must accumulate in the trans-Golgi network at early times and then redistribute to cell junctions to promote cell-cell spread.J Virol. 2006 Apr;80(7):3167-79. doi: 10.1128/JVI.80.7.3167-3179.2006. J Virol. 2006. PMID: 16537585 Free PMC article.
-
Characterization of domains of herpes simplex virus type 1 glycoprotein E involved in Fc binding activity for immunoglobulin G aggregates.J Virol. 1994 Apr;68(4):2478-85. doi: 10.1128/JVI.68.4.2478-2485.1994. J Virol. 1994. PMID: 7511171 Free PMC article.
-
Different receptors binding to distinct interfaces on herpes simplex virus gD can trigger events leading to cell fusion and viral entry.Virology. 2006 Jan 5;344(1):17-24. doi: 10.1016/j.virol.2005.09.016. Virology. 2006. PMID: 16364731 Review.
Cited by
-
Deletion of the first cysteine-rich region of the varicella-zoster virus glycoprotein E ectodomain abolishes the gE and gI interaction and differentially affects cell-cell spread and viral entry.J Virol. 2009 Jan;83(1):228-40. doi: 10.1128/JVI.00913-08. Epub 2008 Oct 22. J Virol. 2009. PMID: 18945783 Free PMC article.
-
Immunization strategies to block the herpes simplex virus type 1 immunoglobulin G Fc receptor.J Virol. 2004 Mar;78(5):2562-71. doi: 10.1128/jvi.78.5.2562-2571.2004. J Virol. 2004. PMID: 14963159 Free PMC article.
-
Responses of herpes simplex virus type 1-infected cells to the presence of extracellular antibodies: gE-dependent glycoprotein capping and enhancement in cell-to-cell spread.J Virol. 2003 Jan;77(1):701-8. doi: 10.1128/jvi.77.1.701-708.2003. J Virol. 2003. PMID: 12477873 Free PMC article.
-
Discovery and Characterization of an Aberrant Small Form of Glycoprotein I of Herpes Simplex Virus Type I in Cell Culture.Microbiol Spectr. 2022 Apr 27;10(2):e0265921. doi: 10.1128/spectrum.02659-21. Epub 2022 Mar 29. Microbiol Spectr. 2022. PMID: 35348373 Free PMC article.
-
Crystal structure of the HSV-1 Fc receptor bound to Fc reveals a mechanism for antibody bipolar bridging.PLoS Biol. 2006 Jun;4(6):e148. doi: 10.1371/journal.pbio.0040148. Epub 2006 May 2. PLoS Biol. 2006. PMID: 16646632 Free PMC article.
References
-
- Basu S, Dubin G, Basu M, Nguyen V, Friedman H M. Characterization of regions of herpes simplex virus type 1 glycoprotein E involved in binding the Fc domain of monomeric IgG and in forming a complex with glycoprotein I. J Immunol. 1995;154:260–267. - PubMed
-
- Basu S, Dubin G, Nagashunmugam T, Basu M, Goldstein L T, Wang L, Weeks B, Friedman H. Mapping regions of herpes simplex virus type 1 glycoprotein I required for formation of the viral Fc receptor for monomeric IgG. J Immunol. 1997;158:209–215. - PubMed
-
- Chapman T L, Heikema A P, Bjorkman P J. The inhibitory receptor LIR-1 uses a common binding interaction to recognize class I MHC molecules and the viral homolog UL18. Immunity. 1999;11:603–614. - PubMed
-
- Chapman T L, You I, Joseph I, Bjorkman P J, Morrison S L, Raghavan M. Characterization of the interaction between the herpes simplex virus type 1 Fc receptor and immunoglobulin G. J Biol Chem. 1999;274:6911–6919. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

