Chromatin-dependent cooperativity between constitutive and inducible activation domains in CREB
- PMID: 11689682
- PMCID: PMC99956
- DOI: 10.1128/MCB.21.23.7892-7900.2001
Chromatin-dependent cooperativity between constitutive and inducible activation domains in CREB
Abstract
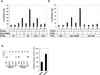
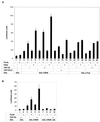
The cyclic AMP (cAMP)-responsive factor CREB induces target gene expression via constitutive (Q2) and inducible (KID, for kinase-inducible domain) activation domains that function synergistically in response to cellular signals. KID stimulates transcription via a phospho (Ser133)-dependent interaction with the coactivator paralogs CREB binding protein and p300, whereas Q2 recruits the TFIID complex via a direct association with hTAF(II)130. Here we investigate the mechanism underlying cooperativity between the Q2 domain and KID in CREB by in vitro transcription assay with naked DNA and chromatin templates containing the cAMP-responsive somatostatin promoter. The Q2 domain was highly active on a naked DNA template, and Ser133 phosphorylation had no additional effect on transcriptional initiation in crude extracts. Q2 activity was repressed on a chromatin template, however, and this repression was relieved by the phospho (Ser133) KID-dependent recruitment of p300 histone acetyltransferase activity to the promoter. In chromatin immunoprecipitation assays of NIH 3T3 cells, cAMP-dependent recruitment of p300 to the somatostatin promoter stimulated acetylation of histone H4. Correspondingly, overexpression of hTAFII130 potentiated CREB activity in cells exposed to cAMP, but had no effect on reporter gene expression in unstimulated cells. We propose that cooperativity between the KID and Q2 domains proceeds via a chromatin-dependent mechanism in which recruitment of p300 facilitates subsequent interaction of CREB with TFIID.
Figures




References
-
- Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell. 2000;103:667–678. - PubMed
-
- Arias J, Alberts A, Brindle P, Claret F, Smeal T, Karin M, Feramisco J, Montminy M. Activation of cAMP and mitogen responsive genes relies on a common nuclear factor. Nature. 1994;370:226–228. - PubMed
-
- Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. - PubMed
-
- Brindle P, Linke S, Montminy M. Analysis of a PK-A dependent activator in CREB reveals a new role for the CREM family of repressors. Nature. 1993;364:821–824. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
