Methylation-mediated proviral silencing is associated with MeCP2 recruitment and localized histone H3 deacetylation
- PMID: 11689684
- PMCID: PMC99960
- DOI: 10.1128/MCB.21.23.7913-7922.2001
Methylation-mediated proviral silencing is associated with MeCP2 recruitment and localized histone H3 deacetylation
Abstract
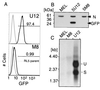
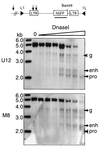
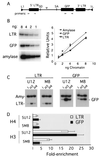
The majority of 5-methylcytosine in mammalian DNA resides in endogenous transposable elements and is associated with the transcriptional silencing of these parasitic elements. Methylation also plays an important role in the silencing of exogenous retroviruses. One of the difficulties inherent in the study of proviral silencing is that the sites in which proviruses randomly integrate influence the probability of de novo methylation and expression. In order to compare methylated and unmethylated proviruses at the same genomic site, we used a recombinase-based targeting approach to introduce an in vitro methylated or unmethylated Moloney murine leukemia-based provirus in MEL cells. The methylated and unmethylated states are maintained in vivo, with the exception of the initially methylated proviral enhancer, which becomes demethylated in vivo. Although the enhancer is unmethylated and remodeled, the methylated provirus is transcriptionally silent. To further analyze the repressed state, histone acetylation status was determined by chromatin immunoprecipitation (ChIP) analyses, which revealed that localized histone H3 but not histone H4 hyperacetylation is inversely correlated with proviral methylation density. Since members of the methyl-CpG binding domain (MBD) family of proteins recruit histone deacetylase activity, these proteins may play a role in proviral repression. Interestingly, only MBD3 and MeCP2 are expressed in MEL cells. ChIPs with antibodies specific for these proteins revealed that only MeCP2 associates with the provirus in a methylation-dependent manner. Taken together, our results suggest that MeCP2 recruitment to a methylated provirus is sufficient for transcriptional silencing, despite the presence of a remodeled enhancer.
Figures




References
-
- Amir R E, Van den Veyver I B, Wan M, Tran C Q, Francke U, Zoghbi H Y. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. - PubMed
-
- Anderson M T, Tjioe I M, Lorincz M C, Parks D R, Herzenberg L A, Nolan G P, Herzenberg L A. Simultaneous fluorescence-activated cell sorter analysis of two distinct transcriptional elements within a single cell using engineered green fluorescent proteins. Proc Natl Acad Sci USA. 1996;93:8508–8511. - PMC - PubMed
-
- Baer A, Schübeler D, Bode J. Transcriptional properties of genomic transgene integration sites marked by electroporation or retroviral infection. Biochemistry. 2000;39:7041–7049. - PubMed
-
- Bestor T H. The host defence function of genomic methylation patterns. Novartis Found Symp. 1998;214:187–195. - PubMed
-
- Bestor T H, Tycko B. Creation of genomic methylation patterns. Nat Genet. 1996;12:363–367. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
