Individual somatic H1 subtypes are dispensable for mouse development even in mice lacking the H1(0) replacement subtype
- PMID: 11689686
- PMCID: PMC99962
- DOI: 10.1128/MCB.21.23.7933-7943.2001
Individual somatic H1 subtypes are dispensable for mouse development even in mice lacking the H1(0) replacement subtype
Abstract
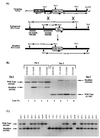
H1 linker histones are involved in facilitating the folding of chromatin into a 30-nm fiber. Mice contain eight H1 subtypes that differ in amino acid sequence and expression during development. Previous work showed that mice lacking H1(0), the most divergent subtype, develop normally. Examination of chromatin in H1(0-/-) mice showed that other H1s, especially H1c, H1d, and H1e, compensate for the loss of H1(0) to maintain a normal H1-to-nucleosome stoichiometry, even in tissues that normally contain abundant amounts of H1(0) (A. M. Sirotkin et al., Proc. Natl. Acad. Sci. USA 92:6434-6438, 1995). To further investigate the in vivo role of individual mammalian H1s in development, we generated mice lacking H1c, H1d, or H1e by homologous recombination in mouse embryonic stem cells. Mice lacking any one of these H1 subtypes grew and reproduced normally and did not exhibit any obvious phenotype. To determine whether one of these H1s, in particular, was responsible for the compensation present in H1(0-/-) mice, each of the three H1 knockout mouse lines was bred with H1(0) knockout mice to generate H1c/H1(0), H1d/H1(0), or H1e/H1(0) double-knockout mice. Each of these doubly H1-deficient mice also was fertile and exhibited no anatomic or histological abnormalities. Chromatin from the three double-knockout strains showed no significant change in the ratio of total H1 to nucleosomes. These results suggest that any individual H1 subtype is dispensable for mouse development and that loss of even two subtypes is tolerated if a normal H1-to-nucleosome stoichiometry is maintained. Multiple compound H1 knockouts will probably be needed to disrupt the compensation within this multigene family.
Figures




References
-
- Brannan C I, Gilbert D J, Ceci J D, Matsuda Y, Chapman V M, Mercer J A, Eisen H, Johnston L A, Copeland N G, Jenkins N A. An interspecific linkage map of mouse chromosome 15 positioned with respect to the centromere. Genomics. 1992;13:1075–1081. - PubMed
-
- Brown D T, Sittman D B. Identification through overexpression and tagging of the variant type of the mouse H1e and H1c genes. J Biol Chem. 1993;268:713–718. - PubMed
-
- Drabent B, Saftig P, Bode C, Doenecke D. Spermatogenesis proceeds normally in mice without linker histone H1t. Histochem Cell Biol. 2000;113:433–442. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
