Construction and analysis of mouse strains lacking the ubiquitin ligase UBR1 (E3alpha) of the N-end rule pathway
- PMID: 11689692
- PMCID: PMC99968
- DOI: 10.1128/MCB.21.23.8007-8021.2001
Construction and analysis of mouse strains lacking the ubiquitin ligase UBR1 (E3alpha) of the N-end rule pathway
Abstract
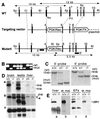
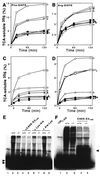
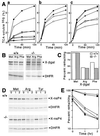
The N-end rule relates the in vivo half-life of a protein to the identity of its N-terminal residue. In the yeast Saccharomyces cerevisiae, the UBR1-encoded ubiquitin ligase (E3) of the N-end rule pathway mediates the targeting of substrate proteins in part through binding to their destabilizing N-terminal residues. The functions of the yeast N-end rule pathway include fidelity of chromosome segregation and the regulation of peptide import. Our previous work described the cloning of cDNA and a gene encoding the 200-kDa mouse UBR1 (E3alpha). Here we show that mouse UBR1, in the presence of a cognate mouse ubiquitin-conjugating (E2) enzyme, can rescue the N-end rule pathway in ubr1Delta S. cerevisiae. We also constructed UBR1(-/-) mouse strains that lacked the UBR1 protein. UBR1(-/-) mice were viable and fertile but weighed significantly less than congenic +/+ mice. The decreased mass of UBR1(-/-) mice stemmed at least in part from smaller amounts of the skeletal muscle and adipose tissues. The skeletal muscle of UBR1(-/-) mice apparently lacked the N-end rule pathway and exhibited abnormal regulation of fatty acid synthase upon starvation. By contrast, and despite the absence of the UBR1 protein, UBR1(-/-) fibroblasts contained the N-end rule pathway. Thus, UBR1(-/-) mice are mosaics in regard to the activity of this pathway, owing to differential expression of proteins that can substitute for the ubiquitin ligase UBR1 (E3alpha). We consider these UBR1-like proteins and discuss the functions of the mammalian N-end rule pathway.
Figures



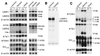
References
-
- Alagramam K, Naider F, Becker J M. A. Recognition component of the ubiquitin system is required for peptide transport in Saccharomyces cerevisiae. Mol Microbiol. 1995;15:225–234. - PubMed
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Wiley-Interscience; 2000.
-
- Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. - PubMed
-
- Bachmair A, Varshavsky A. The degradation signal in a short-lived protein. Cell. 1989;56:1019–1032. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
