The RNA-binding protein Tsunagi interacts with Mago Nashi to establish polarity and localize oskar mRNA during Drosophila oogenesis
- PMID: 11691839
- PMCID: PMC312802
- DOI: 10.1101/gad.927001
The RNA-binding protein Tsunagi interacts with Mago Nashi to establish polarity and localize oskar mRNA during Drosophila oogenesis
Abstract
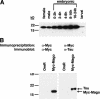
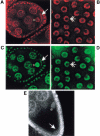
In Drosophila melanogaster, formation of the axes and the primordial germ cells is regulated by interactions between the germ line-derived oocyte and the surrounding somatic follicle cells. This reciprocal signaling results in the asymmetric localization of mRNAs and proteins critical for these oogenic processes. Mago Nashi protein interprets the posterior follicle cell-to-oocyte signal to establish the major axes and to determine the fate of the primordial germ cells. Using the yeast two-hybrid system we have identified an RNA-binding protein, Tsunagi, that interacts with Mago Nashi protein. The proteins coimmunoprecipitate and colocalize, indicating that they form a complex in vivo. Immunolocalization reveals that Tsunagi protein is localized within the posterior oocyte cytoplasm during stages 1-5 and 8-9, and that this localization is dependent on wild-type mago nashi function. When tsunagi function is removed from the germ line, egg chambers develop in which the oocyte nucleus fails to migrate, oskar mRNA is not localized within the posterior pole, and dorsal-ventral pattern abnormalities are observed. These results show that a Mago Nashi-Tsunagi protein complex is required for interpreting the posterior follicle cell-to-oocyte signal to define the major body axes and to localize components necessary for determination of the primordial germ cells.
Figures





Similar articles
-
Mago Nashi and Tsunagi/Y14, respectively, regulate Drosophila germline stem cell differentiation and oocyte specification.Dev Biol. 2007 Aug 15;308(2):507-19. doi: 10.1016/j.ydbio.2007.06.007. Epub 2007 Jun 13. Dev Biol. 2007. PMID: 17628520 Free PMC article.
-
The mago nashi gene is required for the polarisation of the oocyte and the formation of perpendicular axes in Drosophila.Curr Biol. 1997 Jul 1;7(7):468-78. doi: 10.1016/s0960-9822(06)00218-1. Curr Biol. 1997. PMID: 9210377
-
Mago Nashi, Tsunagi/Y14, and Ranshi form a complex that influences oocyte differentiation in Drosophila melanogaster.Dev Biol. 2010 Mar 15;339(2):307-19. doi: 10.1016/j.ydbio.2009.12.035. Epub 2010 Jan 4. Dev Biol. 2010. PMID: 20045686 Free PMC article.
-
Multiple signaling pathways establish both the individuation and the polarity of the oocyte follicle in Drosophila.Arch Insect Biochem Physiol. 1996;33(3-4):211-30. doi: 10.1002/(SICI)1520-6327(1996)33:3/4<211::AID-ARCH4>3.0.CO;2-V. Arch Insect Biochem Physiol. 1996. PMID: 8913032 Review.
-
Axis formation during Drosophila oogenesis.Curr Opin Genet Dev. 2001 Aug;11(4):374-83. doi: 10.1016/s0959-437x(00)00207-0. Curr Opin Genet Dev. 2001. PMID: 11448623 Review.
Cited by
-
A specific set of exon junction complex subunits is required for the nuclear retention of unspliced RNAs in Caenorhabditis elegans.Mol Cell Biol. 2013 Jan;33(2):444-56. doi: 10.1128/MCB.01298-12. Epub 2012 Nov 12. Mol Cell Biol. 2013. PMID: 23149939 Free PMC article.
-
The Physiological Roles of the Exon Junction Complex in Development and Diseases.Cells. 2022 Apr 1;11(7):1192. doi: 10.3390/cells11071192. Cells. 2022. PMID: 35406756 Free PMC article. Review.
-
Sm proteins specify germ cell fate by facilitating oskar mRNA localization.Development. 2010 Jul;137(14):2341-51. doi: 10.1242/dev.042721. Development. 2010. PMID: 20570937 Free PMC article.
-
Ooplasmic flow cooperates with transport and anchorage in Drosophila oocyte posterior determination.J Cell Biol. 2018 Oct 1;217(10):3497-3511. doi: 10.1083/jcb.201709174. Epub 2018 Jul 23. J Cell Biol. 2018. PMID: 30037924 Free PMC article.
-
Nuclear RNA export factor 7 is localized in processing bodies and neuronal RNA granules through interactions with shuttling hnRNPs.Nucleic Acids Res. 2008 Feb;36(2):616-28. doi: 10.1093/nar/gkm556. Epub 2007 Dec 5. Nucleic Acids Res. 2008. PMID: 18063567 Free PMC article.
References
-
- Bashirullah A, Cooperstock RL, Lipshitz HD. RNA localization in development. Ann Rev Biochem. 1998;67:335–394. - PubMed
-
- Boswell RE, Prout ME, Steichen JC. Mutations in a newly identified Drosophila melanogaster gene, mago nashi, disrupt germ cell formation and result in the formation of mirror-image symmetrical double abdomen embryos. Development. 1991;113:373–384. - PubMed
-
- Bousquet-Antonelli C, Presutti C, Tollervey D. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell. 2000;102:765–775. - PubMed
-
- Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
