A short pseudoautosomal region in laboratory mice
- PMID: 11691846
- PMCID: PMC311143
- DOI: 10.1101/gr.203001
A short pseudoautosomal region in laboratory mice
Abstract
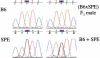
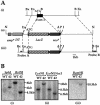
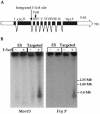
The pseudoautosomal region (PAR) of mammalian sex chromosomes is a small region of sequence identity that is the site of an obligatory pairing and recombination event between the X and Y chromosomes during male meiosis. During female meiosis, X chromosomes can pair and recombine along their entire length; recombination in the PAR is therefore approximately 10x greater in male meiosis compared with female meiosis. A consequence of the presence of the PAR in two copies in males and females is that genes in the region escape the process of X-inactivation. Although the structure and gene content of the human PAR at Xq/Yq is well understood, the mouse PAR, which appears to be of independent evolutionary origin, is poorly characterized. Here we describe a yeast artificial chromosome (YAC) contig covering the distal part of the mouse X chromosome, which we have used to define the pseudoautosomal boundary, that is, the point of divergence of X-specific and X-Y-identical sequences. In addition, we have investigated the size of the mouse PAR by integrating a unique restriction endonuclease recognition site just proximal to the pseudoautosomal boundary by homologous recombination. Restriction digestion of this modified DNA and pulsed field gel electrophoresis reveal that the PAR in these cells is approximately 700 kb. Thus, the mouse PAR, although small in size, has retained essential sex chromosome pairing functions despite its rapid rate of evolution.
Figures



References
-
- Birren B, Lai E. Pulsed field gel electrophoresis: A practical guide. San Diego: Academic Press; 1993.
-
- Burgoyne PS. Genetic homology and crossing over in the X and Y chromosomes of mammals. Hum Genet. 1982;61:85–90. - PubMed
-
- Burgoyne PS, Mahadevaiah SK, Sutcliffe MJ, Palmer SJ. Fertility in mice requires X-Y pairing and a Y-chromosomal “spermiogenesis” gene mapping to the long arm. Cell. 1992;71:391–398. - PubMed
-
- Ciccodicola A, D'Esposito M, Esposito T, Gianfrancesco F, Migliaccio C, Miano MG, Matarazzo MR, Vacca M, Franze A, Cuccurese M, et al. Differentially regulated and evolved genes in the fully sequenced Xq/Yq pseudoautosomal region. Hum Mol Genet. 2000;9:395–401. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
