The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes
- PMID: 11691919
- PMCID: PMC60187
- DOI: 10.1093/nar/29.21.4319
The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes
Abstract
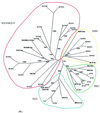
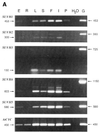
SET domains are conserved amino acid motifs present in chromosomal proteins that function in epigenetic control of gene expression. These proteins can be divided into four classes as typified by their Drosophila members E(Z), TRX, ASH1 and SU(VAR)3-9. Homologs of all four classes have been identified in yeast and mammals, but not in plants. A BLASTP screening of the Arabidopsis genome identified 37 genes: three E(z) homologs, five trx homologs, four ash1 homologs and 15 genes similar to Su(var)3-9. Seven genes were assigned as trx-related and three as ash1-related. Only four genes have been described previously. Our classification is based on the characteristics of the SET domains, cysteine-rich regions and additional conserved domains, including a novel YGD domain. RT-PCR analysis, cDNA cloning and matching ESTs show that at least 29 of the genes are active in diverse tissues. The high number of SET domain genes, possibly involved in epigenetic control of gene activity during plant development, can partly be explained by extensive genome duplication in Arabidopsis. Additionally, the lack of introns in the coding region of eight SU(VAR)3-9 class genes indicates evolution of new genes by retrotransposition. The identification of putative nuclear localization signals and AT-hooks in many of the proteins supports an anticipated nuclear localization, which was demonstrated for selected proteins.
Figures





References
-
- Henikoff S. (1996) Position-effect variegation in Drosophila. In Russo,V.E.A. (ed.), Epigenetic Mechanisms of Gene Regulation. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 319–334.
-
- Wallrath L. (1998) Unfolding the mysteries of heterochromatin. Curr. Opin. Genet. Dev., 8, 147–153. - PubMed
-
- Weiler K.S. and Wakimoto,B.T. (1995) Heterochromatin and gene expression in Drosophila. Annu. Rev. Genet., 29, 577–605. - PubMed
-
- Aagaard L., Laible,G., Selenko,P., Schmid,M., Dorn,R., Schotta,G., Kuhfittig,S., Wolf,A., Lebersorger,A., Singh,P.B. et al. (1999) Functional mammalian homologs of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J., 18, 1923–1938. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

