Completion of replication map of Saccharomyces cerevisiae chromosome III
- PMID: 11694569
- PMCID: PMC60257
- DOI: 10.1091/mbc.12.11.3317
Completion of replication map of Saccharomyces cerevisiae chromosome III
Abstract
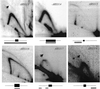
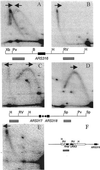
In Saccharomyces cerevisiae chromosomal DNA replication initiates at intervals of approximately 40 kb and depends upon the activity of autonomously replicating sequence (ARS) elements. The identification of ARS elements and analysis of their function as chromosomal replication origins requires the use of functional assays because they are not sufficiently similar to identify by DNA sequence analysis. To complete the systematic identification of ARS elements on S. cerevisiae chromosome III, overlapping clones covering 140 kb of the right arm were tested for their ability to promote extrachromosomal maintenance of plasmids. Examination of chromosomal replication intermediates of each of the seven ARS elements identified revealed that their efficiencies of use as chromosomal replication origins varied widely, with four ARS elements active in < or = 10% of cells in the population and two ARS elements active in > or = 90% of the population. Together with our previous analysis of a 200-kb region of chromosome III, these data provide the first complete analysis of ARS elements and DNA replication origins on an entire eukaryotic chromosome.
Figures



References
-
- Abraham J, Nasmyth KA, Strathern JN, Klar AJ, Hicks JB. Regulation of mating-type information in yeast. Negative control requiring sequences both 5′ and 3′ to the regulated region. J Mol Biol. 1984;176:307–331. - PubMed
-
- Boyer HW, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. - PubMed
-
- Brewer BJ, Fangman WL. Initiation at closely spaced replication origins in a yeast chromosome. Science. 1993;262:1728–1731. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

