Functions of the mismatch repair gene mutS from Acinetobacter sp. strain ADP1
- PMID: 11698371
- PMCID: PMC95523
- DOI: 10.1128/JB.183.23.6822-6831.2001
Functions of the mismatch repair gene mutS from Acinetobacter sp. strain ADP1
Abstract
The genus Acinetobacter encompasses a heterogeneous group of bacteria that are ubiquitous in the natural environment due in part to their ability to adapt genetically to novel challenges. Acinetobacter sp. strain ADP1 (also known as strain BD413) is naturally transformable and takes up DNA from any source. Donor DNA can be integrated into the chromosome by recombination provided it possesses sufficient levels of nucleotide sequence identity to the recipient's DNA. In other bacteria, the requirement for sequence identity during recombination is partly due to the actions of the mismatch repair system, a key component of which, MutS, recognizes mismatched bases in heteroduplex DNA and, along with MutL, blocks strand exchange. We have cloned mutS from strain ADP1 and examined its roles in preventing recombination between divergent DNA and in the repair of spontaneous replication errors. Inactivation of mutS resulted in 3- to 17-fold increases in transformation efficiencies with donor sequences that were 8 to 20% divergent relative to the strain ADP1. Strains lacking MutS exhibited increased spontaneous mutation frequencies, and reversion assays demonstrated that MutS preferentially recognized transition mismatches while having little effect on the repair of transversion mismatches. Inactivation of mutS also abolished the marker-specific variations in transforming efficiency seen in mutS(+) recipients where transition and frameshift alleles transformed at eightfold lower frequencies than transversions or large deletions. Comparison of the MutS homologs from five individual Acinetobacter strains with those of other gram-negative bacteria revealed that a number of unique indels are conserved among the Acinetobacter amino acid sequences.
Figures
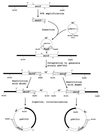

 represents
putative transcription terminators downstream from mutS
(5′-ATAAGTAGCCATCGTGCTACTTAT-3′) and downstream from
fdxA (5′-AAAAGATCAGCATTAGCTGATCTTTT-3′).
Horizontal lines indicate inserts in plasmids containing
overlapping portions of mutS.
represents
putative transcription terminators downstream from mutS
(5′-ATAAGTAGCCATCGTGCTACTTAT-3′) and downstream from
fdxA (5′-AAAAGATCAGCATTAGCTGATCTTTT-3′).
Horizontal lines indicate inserts in plasmids containing
overlapping portions of mutS.


References
-
- Cox E C. Bacterial mutator genes and the control of spontaneous mutation. Annu Rev Genet. 1976;10:135–156. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

