Binding site of macrolide antibiotics on the ribosome: new resistance mutation identifies a specific interaction of ketolides with rRNA
- PMID: 11698379
- PMCID: PMC95531
- DOI: 10.1128/JB.183.23.6898-6907.2001
Binding site of macrolide antibiotics on the ribosome: new resistance mutation identifies a specific interaction of ketolides with rRNA
Abstract
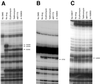
Macrolides represent a clinically important class of antibiotics that block protein synthesis by interacting with the large ribosomal subunit. The macrolide binding site is composed primarily of rRNA. However, the mode of interaction of macrolides with rRNA and the exact location of the drug binding site have yet to be described. A new class of macrolide antibiotics, known as ketolides, show improved activity against organisms that have developed resistance to previously used macrolides. The biochemical reasons for increased potency of ketolides remain unknown. Here we describe the first mutation that confers resistance to ketolide antibiotics while leaving cells sensitive to other types of macrolides. A transition of U to C at position 2609 of 23S rRNA rendered E. coli cells resistant to two different types of ketolides, telithromycin and ABT-773, but increased slightly the sensitivity to erythromycin, azithromycin, and a cladinose-containing derivative of telithromycin. Ribosomes isolated from the mutant cells had reduced affinity for ketolides, while their affinity for erythromycin was not diminished. Possible direct interaction of ketolides with position 2609 in 23S rRNA was further confirmed by RNA footprinting. The newly isolated ketolide-resistance mutation, as well as 23S rRNA positions shown previously to be involved in interaction with macrolide antibiotics, have been modeled in the crystallographic structure of the large ribosomal subunit. The location of the macrolide binding site in the nascent peptide exit tunnel at some distance from the peptidyl transferase center agrees with the proposed model of macrolide inhibitory action and explains the dominant nature of macrolide resistance mutations. Spatial separation of the rRNA residues involved in universal contacts with macrolides from those believed to participate in structure-specific interactions with ketolides provides the structural basis for the improved activity of the broader spectrum group of macrolide antibiotics.
Figures
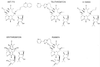



References
-
- Arevalo M A, Tejedor F, Polo F, Ballesta J P. Protein components of the erythromycin binding site in bacterial ribosomes. J Biol Chem. 1988;263:58–63. - PubMed
-
- Ban N, Nissen P, Hansen J, Moore P B, Steitz T A. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. - PubMed
-
- Beauclerk A A, Cundliffe E. Sites of action of two ribosomal RNA methylases responsible for resistance to aminoglycosides. J Mol Biol. 1987;193:661–671. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

