Extraordinary variation in a diversified family of immune-type receptor genes
- PMID: 11698645
- PMCID: PMC61127
- DOI: 10.1073/pnas.231418598
Extraordinary variation in a diversified family of immune-type receptor genes
Abstract
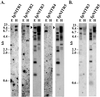
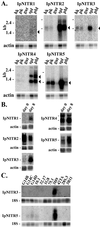
Immune inhibitory receptor genes that encode a variable (V) region, a unique V-like C2 (V/C2) domain, a transmembrane region, and a cytoplasmic tail containing immunoreceptor tyrosine-based inhibition motifs (ITIMs) have been described previously in two lineages of bony fish. In the present study, eleven related genes encoding distinct structural forms have been identified in Ictalurus punctatus (channel catfish), a well characterized immunological model system that represents a third independent bony fish lineage. Each of the different genes encodes an N-terminal V region but differs in the number of extracellular Ig domains, number and location of joining (J) region-like motifs, presence of transmembrane regions, presence of charged residues in transmembrane regions, presence of cytoplasmic tails, and/or distribution of ITIM(s) within the cytoplasmic tails. Variation in the numbers of genomic copies of the different gene types, their patterns of expression, and relative levels of expression in mixed leukocyte cultures (MLC) is reported. V region-containing immune-type genes constitute a far more complex family than recognized originally and include individual members that might function in inhibitory or, potentially activatory manners.
Figures



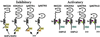
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

