Cholesterol depletion induces large scale domain segregation in living cell membranes
- PMID: 11698680
- PMCID: PMC60826
- DOI: 10.1073/pnas.231377398
Cholesterol depletion induces large scale domain segregation in living cell membranes
Abstract
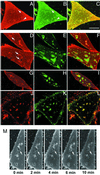
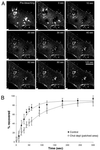
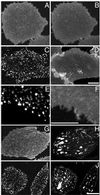
Local inhomogeneities in lipid composition play a crucial role in regulation of signal transduction and membrane traffic. Nevertheless, most evidence for microdomains in cells remains indirect, and the nature of membrane inhomogeneities has been difficult to characterize. We used lipid analogs and lipid-anchored proteins with varying fluidity preferences to examine the effect of modulating cellular cholesterol on domain formation. We show that lowering cholesterol levels induces formation of visible micrometer-scale domains in the plasma membrane of several mammalian cell types with complementary distributions of fluorescent lipid analogs with preferences for fluid or ordered domains. A uniform distribution is restored by cholesterol repletion. Unexpectedly, cholesterol depletion does not visibly alter the distribution of a crosslinked or uncrosslinked glycosylphosphatidylinositol-anchored protein (the folate receptor). We also examined the effect of varying cholesterol content on the cold Triton X-100 solubility of several membrane constituents. Although a cholesterol analog, dehydroergosterol, and a glycosylphosphatidylinositol-anchored protein are largely retained after extraction, a lipid analog with saturated 16-carbon acyl chains is largely removed when the cellular cholesterol level is lowered. This result indicates that after cholesterol depletion molecules in the more ordered domains can be extracted differentially by cold nonionic detergents.
Figures

References
-
- Brown D A, London E. J Biol Chem. 2000;275:17221–17224. - PubMed
-
- Simons K, Ikonen E. Nature (London) 1997;387:569–572. - PubMed
-
- Brown D A, Rose J K. Cell. 1992;68:533–544. - PubMed
-
- Brown D A, London E. J Membr Biol. 1998;164:103–114. - PubMed
-
- Roper K, Corbeil D, Huttner W B. Nat Cell Biol. 2000;2:582–592. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

