The Arabidopsis BELL1 and KNOX TALE homeodomain proteins interact through a domain conserved between plants and animals
- PMID: 11701881
- PMCID: PMC139464
- DOI: 10.1105/tpc.010161
The Arabidopsis BELL1 and KNOX TALE homeodomain proteins interact through a domain conserved between plants and animals
Abstract
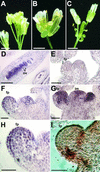
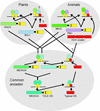
Interactions between TALE (three-amino acid loop extension) homeodomain proteins play important roles in the development of both fungi and animals. Although in plants, two different subclasses of TALE proteins include important developmental regulators, the existence of interactions between plant TALE proteins has remained unexplored. We have used the yeast two-hybrid system to demonstrate that the Arabidopsis BELL1 (BEL1) homeodomain protein can selectively heterodimerize with specific KNAT homeodomain proteins. Interaction is mediated by BEL1 sequences N terminal to the homeodomain and KNAT sequences including the MEINOX domain. These findings validate the hypothesis that the MEINOX domain has been conserved between plants and animals as an interaction domain for developmental regulators. In yeast, BEL1 and KNAT proteins can activate transcription only as a heterodimeric complex, suggesting a role for such complexes in planta. Finally, overlapping patterns of BEL1 and SHOOT MERISTEMLESS (STM) expression within the inflorescence meristem suggest a role for the BEL1-STM complex in maintaining the indeterminacy of the inflorescence meristem.
Figures




References
-
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815. - PubMed
-
- Bartel, P., Chien, C.T., Sternglanz, R., and Fields, S. (1993). Elimination of false positives that arise in using the 2-hybrid system. Biotechniques 14 920–924. - PubMed
-
- Barton, M.K., and Poethig, R.S. (1993). Formation of the shoot apical meristem in Arabidopsis thaliana: An analysis of the development in the wild-type and shoot meristemless mutant. Development 119 823–831.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

