Evidence that mycobacterial PE_PGRS proteins are cell surface constituents that influence interactions with other cells
- PMID: 11705904
- PMCID: PMC98818
- DOI: 10.1128/IAI.69.12.7326-7333.2001
Evidence that mycobacterial PE_PGRS proteins are cell surface constituents that influence interactions with other cells
Abstract
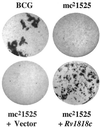
The elucidation of the genomic sequence of Mycobacterium tuberculosis revealed the presence of a novel multigene family designated PE/PE_PGRS that encodes numerous, highly related proteins of unknown function. In this study, we demonstrate that a transposon insertion in a PE_PGRS gene (1818(PE_PGRS)) found in Mycobacterium bovis BCG Pasteur, which is the BCG homologue of the M. tuberculosis H37Rv gene Rv1818c, introduces new phenotypic properties to this BCG strain. These properties include dispersed growth in liquid medium and reduced infection of macrophages. Complementation of the 1818(PE_PGRS)::Tn5367 mutant with the wild-type gene restores both aggregative growth (clumping) in liquid medium and reestablishes infectivity of macrophages to levels equivalent to those for the parent BCG strain. Western blot analysis using antisera raised against the 1818(PE_PGRS) protein shows that PE_PGRS proteins are found in cell lysates of BCG and M. tuberculosis H37Ra and in the cell wall fraction of M. tuberculosis H37Rv. Moreover, immunofluorescent labeling of mycobacteria indicates that certain PE_PGRS proteins are localized at the cell surface of BCG and M. tuberculosis. Together these results suggest that certain PE_PGRS proteins may be found at the surface of mycobacteria and influence both cell surface interactions among mycobacteria as well as the interactions of mycobacteria with macrophages.
Figures




References
-
- Abraham S N, Jonsson A B, Normark S. Fimbriae-mediated host-pathogen cross-talk. Curr Opin Microbiol. 1998;1:75–81. - PubMed
-
- Arruda S, Bomfim G, Knights R, Huima-Byron T, Riley L W. Cloning of an M. tuberculosis DNA fragment associated with entry and survival inside cells. Science. 1993;261:1454–1457. - PubMed
-
- Behr M A, Wilson M A, Gill W P, Salamon H, Schoolnik G K, Rane S, Small P M. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science. 1999;284:1520–1523. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

