Identification, characterization, and variable expression of a naturally occurring inhibitor protein of IS1106 transposase in clinical isolates of Neisseria meningitidis
- PMID: 11705917
- PMCID: PMC98831
- DOI: 10.1128/IAI.69.12.7425-7436.2001
Identification, characterization, and variable expression of a naturally occurring inhibitor protein of IS1106 transposase in clinical isolates of Neisseria meningitidis
Abstract
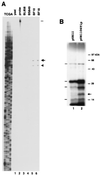
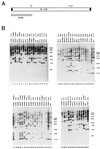
Transposition plays a role in the epidemiology and pathogenesis of Neisseria meningitidis. Insertion sequences are involved in reversible capsulation and insertional inactivation of virulence genes encoding outer membrane proteins. In this study, we have investigated and identified one way in which transposon IS1106 controls its own activity. We have characterized a naturally occurring protein (Tip) that inhibits the transposase. The inhibitor protein is a truncated version of the IS1106 transposase lacking the NH(2)-terminal DNA binding sequence, and it regulates transposition by competing with the transposase for binding to the outside ends of IS1106, as shown by gel shift and in vitro transposition assays. IS1106Tip mRNA is variably expressed among serogroup B meningococcal clinical isolates, and it is absent in most collection strains belonging to hypervirulent lineages.
Figures





Similar articles
-
Genetic analysis of Canadian isolates of C:2a:P1.2,5 and B:2a:P1.2,5 Neisseria meningitidis strains belonging to the hypervirulent clone of ET-15.Can J Microbiol. 2004 Jun;50(6):433-43. doi: 10.1139/w04-024. Can J Microbiol. 2004. PMID: 15284889
-
Sequencing of porA from clinical isolates of Neisseria meningitidis defines a subtyping scheme and its genetic regulation.Can J Microbiol. 1998 Jan;44(1):56-63. Can J Microbiol. 1998. PMID: 9522450
-
Genetic mechanisms for loss of encapsulation in polysialyltransferase-gene-positive meningococci isolated from healthy carriers.Int J Med Microbiol. 2006 Nov;296(7):475-84. doi: 10.1016/j.ijmm.2006.05.004. Epub 2006 Jul 28. Int J Med Microbiol. 2006. PMID: 16876478
-
Assembly of the Tc1 and mariner transposition initiation complexes depends on the origins of their transposase DNA binding domains.Genetica. 2007 Jun;130(2):105-20. doi: 10.1007/s10709-006-0025-2. Epub 2006 Aug 16. Genetica. 2007. PMID: 16912840 Review.
-
Intermediate molecules generated by transposase in the pathways of transposition of bacterial insertion element IS3.Adv Biophys. 2004;38:125-39. Adv Biophys. 2004. PMID: 15493331 Review. No abstract available.
Cited by
-
Phenotypes of a naturally defective recB allele in Neisseria meningitidis clinical isolates.Infect Immun. 2002 Aug;70(8):4185-95. doi: 10.1128/IAI.70.8.4185-4195.2002. Infect Immun. 2002. PMID: 12117927 Free PMC article.
-
Bacterial insertion sequences: their genomic impact and diversity.FEMS Microbiol Rev. 2014 Sep;38(5):865-91. doi: 10.1111/1574-6976.12067. Epub 2014 Feb 26. FEMS Microbiol Rev. 2014. PMID: 24499397 Free PMC article. Review.
-
HrpA anchors meningococci to the dynein motor and affects the balance between apoptosis and pyroptosis.J Biomed Sci. 2022 Jun 28;29(1):45. doi: 10.1186/s12929-022-00829-8. J Biomed Sci. 2022. PMID: 35765029 Free PMC article.
-
Serogroup-specific interaction of Neisseria meningitidis capsular polysaccharide with host cell microtubules and effects on tubulin polymerization.Infect Immun. 2014 Jan;82(1):265-74. doi: 10.1128/IAI.00501-13. Epub 2013 Oct 28. Infect Immun. 2014. PMID: 24166951 Free PMC article.
-
Identification of a meningococcal L-glutamate ABC transporter operon essential for growth in low-sodium environments.Infect Immun. 2006 Mar;74(3):1725-40. doi: 10.1128/IAI.74.3.1725-1740.2006. Infect Immun. 2006. PMID: 16495545 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

