Identification of the sigB operon in Staphylococcus epidermidis: construction and characterization of a sigB deletion mutant
- PMID: 11705980
- PMCID: PMC98894
- DOI: 10.1128/IAI.69.12.7933-7936.2001
Identification of the sigB operon in Staphylococcus epidermidis: construction and characterization of a sigB deletion mutant
Abstract
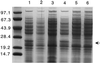
The role of the alternative sigma factor sigma(B) in Staphylococcus epidermidis was investigated by the construction, complementation, and characterization of a sigB deletion mutant. Electrophoretic analyses confirmed a profound influence of sigma(B) on the expression of exoproteins and cytoplasmic proteins. Detailed investigation revealed reduced lipase and enhanced protease activity in the sigma(B) mutant. Furthermore, no significant influence of sigma(B) on heterologous biofilm formation or on the activity of the global regulator agr was detected.
Figures



References
-
- Augustin J, Götz F. Transformation of Staphylococcus epidermidis and other staphylococcal species with plasmid DNA by electroporation. FEMS Microbiol Lett. 1990;54:203–207. - PubMed
-
- Brückner R. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol Lett. 1997;151:1–8. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

