The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture
- PMID: 11706164
- PMCID: PMC129253
The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture
Erratum in
- Plant Physiol 2002 Jan;128(1):314
Abstract
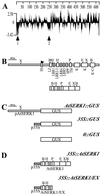
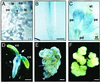
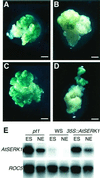
We report here the isolation of the Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE 1 (AtSERK1) gene and we demonstrate its role during establishment of somatic embryogenesis in culture. The AtSERK1 gene is highly expressed during embryogenic cell formation in culture and during early embryogenesis. The AtSERK1 gene is first expressed in planta during megasporogenesis in the nucellus [corrected] of developing ovules, in the functional megaspore, and in all cells of the embryo sac up to fertilization. After fertilization, AtSERK1 expression is seen in all cells of the developing embryo until the heart stage. After this stage, AtSERK1 expression is no longer detectable in the embryo or in any part of the developing seed. Low expression is detected in adult vascular tissue. Ectopic expression of the full-length AtSERK1 cDNA under the control of the cauliflower mosaic virus 35S promoter did not result in any altered plant phenotype. However, seedlings that overexpressed the AtSERK1 mRNA exhibited a 3- to 4-fold increase in efficiency for initiation of somatic embryogenesis. Thus, an increased AtSERK1 level is sufficient to confer embryogenic competence in culture.
Figures


References
-
- Albrecht C, Geurts R, Lapeyrie F, Bisseling T. Endomycorrhizae and rhizobial Nod factors both require SYM8 to induce the expression of the early nodulin genes PsENOD5 and PsENOD12A. Plant J. 1998;15:605–614. - PubMed
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Baudino S, Hansen S, Brettschneider R, Hecht V, Dresselhaus T, Lörz H, Dumas C, Rogowsky P. Molecular characterization of two novel maize LRR receptor-like kinases, which belong to the SERK gene family. Planta. 2001;213:1–10. - PubMed
-
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium mediated transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Paris. 1993;316:1194–1199.
-
- Becker D, Kemper E, Schell J, Masterson R. New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol Biol. 1992;20:1195–1197. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
