Significant accumulation of C(4)-specific pyruvate, orthophosphate dikinase in a C(3) plant, rice
- PMID: 11706193
- PMCID: PMC129282
Significant accumulation of C(4)-specific pyruvate, orthophosphate dikinase in a C(3) plant, rice
Abstract
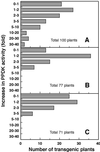
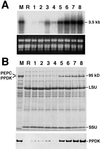
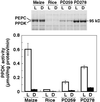
The C(4)-Pdk gene encoding the C(4) enzyme pyruvate, orthophosphate dikinase (PPDK) of maize (Zea mays cv Golden Cross Bantam) was introduced into the C(3) plant, rice (Oryza sativa cv Kitaake). When the intact maize C(4)-Pdk gene, containing its own promoter and terminator sequences and exon/intron structure, was introduced, the PPDK activity in the leaves of some transgenic lines was greatly increased, in one line reaching 40-fold over that of wild-type plants. In a homozygous line, the PPDK protein accounted for 35% of total leaf-soluble protein or 16% of total leaf nitrogen. In contrast, introduction of a chimeric gene containing the full-length cDNA of the maize PPDK fused to the maize C(4)-Pdk promoter or the rice Cab promoter only increased PPDK activity and protein level slightly. These observations suggest that the intron(s) or the terminator sequence of the maize gene, or a combination of both, is necessary for high-level expression. In maize and transgenic rice plants carrying the intact maize gene, the level of transcript in the leaves per copy of the maize C(4)-Pdk gene was comparable, and the maize gene was expressed in a similar organ-specific manner. These results suggest that the maize C(4)-Pdk gene behaves in a quantitatively and qualitatively similar way in maize and transgenic rice plants. The activity of the maize PPDK protein expressed in rice leaves was light/dark regulated as it is in maize. This is the first reported evidence for the presence of an endogenous PPDK regulatory protein in a C(3) plant.
Figures





References
-
- Ashton AR, Burnell JN, Furbank RT, Jenkins CLD, Hatch MD. Enzymes of C4 photosynthesis. In: Lea PJ, editor. Methods in Plant Biochemistry. 3. Enzymes of Primary Metabolism. London: Academic Press; 1990. pp. 39–72.
-
- Bendich AJ. Why do chloroplasts and mitochondria contain so many copies of their genome? BioEssays. 1987;6:279–282. - PubMed
-
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
