Characterization of a tomato xyloglucan endotransglycosylase gene that is down-regulated by auxin in etiolated hypocotyls
- PMID: 11706197
- PMCID: PMC129286
- DOI: 10.1104/pp.010481
Characterization of a tomato xyloglucan endotransglycosylase gene that is down-regulated by auxin in etiolated hypocotyls
Abstract
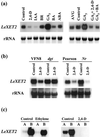
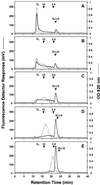
The reorganization of the cellulose-xyloglucan matrix is proposed to serve as an important mechanism in the control of strength and extensibility of the plant primary cell wall. One of the key enzymes associated with xyloglucan metabolism is xyloglucan endotransglycosylase (XET), which catalyzes the endocleavage and religation of xyloglucan molecules. As with other plant species, XETs are encoded by a gene family in tomato (Lycopersicon esculentum cv T5). In a previous study, we demonstrated that the tomato XET gene LeEXT was abundantly expressed in the rapidly expanding region of the etiolated hypocotyl and was induced to higher levels by auxin. Here, we report the identification of a new tomato XET gene, LeXET2, that shows a different spatial expression and diametrically opposite pattern of auxin regulation from LeEXT. LeXET2 was expressed more abundantly in the mature nonelongating regions of the hypocotyl, and its mRNA abundance decreased dramatically following auxin treatment of etiolated hypocotyl segments. Analysis of the effect of several plant hormones on LeXET2 expression revealed that the inhibition of LeXET2 mRNA accumulation also occurred with cytokinin treatment. LeXET2 mRNA levels increased significantly in hypocotyl segments treated with gibberellin, but this increase could be prevented by adding auxin or cytokinin to the incubation media. Recombinant LeXET2 protein obtained by heterologous expression in Pichia pastoris exhibited greater XET activity against xyloglucan from tomato than that from three other species. The opposite patterns of expression and differential auxin regulation of LeXET2 and LeEXT suggest that they encode XETs with distinct roles during plant growth and development.
Figures






References
-
- Arrowsmith DA, de Silva J. Characterization of two tomato fruit-expressed cDNAs encoding xyloglucan endotransglycosylases. Plant Mol Biol. 1995;28:391–403. - PubMed
-
- Barrachina C, Lorences EP. Xyloglucan endotransglycosylase activity in pine hypocotyls: intracellular localization and relationship with endogenous growth. Physiol Plant. 1998;102:55–60. - PubMed
-
- Campbell P, Braam J. In vitro activities of four xyloglucan endotransglycosylases from Arabidopsis. Plant J. 1999a;18:371–382. - PubMed
-
- Campbell P, Braam J. Xyloglucan endotransglycosylases: diversity of genes, enzymes and potential wall-modifying functions. Trends Plant Sci. 1999b;4:361–366. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials

