Biochemical characterization of the Arabidopsis biotin synthase reaction. The importance of mitochondria in biotin synthesis
- PMID: 11706201
- PMCID: PMC129290
Biochemical characterization of the Arabidopsis biotin synthase reaction. The importance of mitochondria in biotin synthesis
Abstract
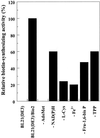
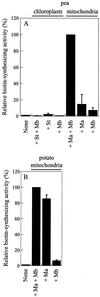
Biotin synthase, encoded by the bio2 gene in Arabidopsis, catalyzes the final step in the biotin biosynthetic pathway. The development of radiochemical and biological detection methods allowed the first detection and accurate quantification of a plant biotin synthase activity, using protein extracts from bacteria overexpressing the Arabidopsis Bio2 protein. Under optimized conditions, the turnover number of the reaction was >2 h(-1) with this in vitro system. Purified Bio2 protein was not efficient by itself in supporting biotin synthesis. However, heterologous interactions between the plant Bio2 protein and bacterial accessory proteins yielded a functional biotin synthase complex. Biotin synthase in this heterologous system obeyed Michaelis-Menten kinetics with respect to dethiobiotin (K(m) = 30 microM) and exhibited a kinetic cooperativity with respect to S-adenosyl-methionine (Hill coefficient = 1.9; K(0.5) = 39 microM), an obligatory cofactor of the reaction. In vitro inhibition of biotin synthase activity by acidomycin, a structural analog of biotin, showed that biotin synthase reaction was the specific target of this inhibitor of biotin synthesis. It is important that combination experiments using purified Bio2 protein and extracts from pea (Pisum sativum) leaf or potato (Solanum tuberosum) organelles showed that only mitochondrial fractions could elicit biotin formation in the plant-reconstituted system. Our data demonstrated that one or more unidentified factors from mitochondrial matrix (pea and potato) and from mitochondrial membranes (pea), in addition to the Bio2 protein, are obligatory for the conversion of dethiobiotin to biotin, highlighting the importance of mitochondria in plant biotin synthesis.
Figures





References
-
- Alban C. Is plant holocarboxylase synthetase a bifunctional enzyme? C R Acad Sci Paris. 2000;323:681–688. - PubMed
-
- Alban C, Job D, Douce R. Biotin metabolism in plants. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:17–47. - PubMed
-
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. - PubMed
-
- Baldet P. Quelques observations sur le métabolisme de la biotine chez les plantes supérieures. PhD thesis. France: University of Grenoble; 1993.
-
- Baldet P, Alban C, Axiotis S, Douce R. Localization of free and bound biotin in cells from green pea leaves. Arch Biochem Biophys. 1993a;303:67–73. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
