Stepwise activation of the immunoglobulin mu heavy chain gene locus
- PMID: 11707410
- PMCID: PMC125735
- DOI: 10.1093/emboj/20.22.6394
Stepwise activation of the immunoglobulin mu heavy chain gene locus
Abstract
The immunoglobulin heavy chain (IgH) gene locus spans several megabases. We show that IgH activation during B-cell differentiation, as measured by histone acetylation, occurs in discrete, independently regulated domains. Initially, a 120 kb domain of germline DNA is hyperacetylated, that extends from D(FL16.1), the 5'-most D(H) gene segment, to the intergenic region between Cmu and Cdelta. Germline V(H) genes were not hyperacetylated at this stage, which accounts for D(H) to J(H) recombination occurring first during B-cell development. Subsequent activation of the V(H) locus happens in at least three differentially regulated domains: an interleukin-7-regulated domain consisting of the 5' J558 family, an intermediate domain and the 3' V(H) genes, which are hyperacetylated in response to DJ(H) recombination. These observations lead to mechanisms for two well-documented phenomena in B-cell ontogeny: the sequential rearrangement of D(H) followed by V(H) gene segments, and the preferential recombination of D(H)-proximal V(H) genes in pro-B cells. We suggest that stepwise activation may be a general mechanism by which large segments of the genome are prepared for expression.
Figures

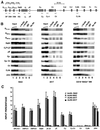
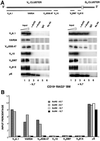
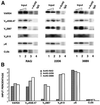
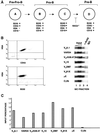
References
-
- Akashi K., Kondo,M. and Weissman,I.L. (1998) Role of interleukin-7 in T-cell development from hematopoietic stem cells. Immunol. Rev., 165, 13–28. - PubMed
-
- Banerjee A. and Rothman,P. (1998) IL-7 reconstitutes multiple aspects of v-Abl-mediated signaling. J. Immunol., 161, 4611–4617. - PubMed
-
- Braunstein M., Rose,A.B., Holmes,S.G., Allis,C.D. and Broach,J.R. (1993) Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev., 7, 592–604. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

