Diversity of gene expression in adenocarcinoma of the lung
- PMID: 11707590
- PMCID: PMC61119
- DOI: 10.1073/pnas.241500798
Diversity of gene expression in adenocarcinoma of the lung
Erratum in
- Proc Natl Acad Sci U S A 2002 Jan 22;99(2):1098
Abstract
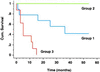
The global gene expression profiles for 67 human lung tumors representing 56 patients were examined by using 24,000-element cDNA microarrays. Subdivision of the tumors based on gene expression patterns faithfully recapitulated morphological classification of the tumors into squamous, large cell, small cell, and adenocarcinoma. The gene expression patterns made possible the subclassification of adenocarcinoma into subgroups that correlated with the degree of tumor differentiation as well as patient survival. Gene expression analysis thus promises to extend and refine standard pathologic analysis.
Figures




References
-
- Travis W, Colby T, Corrin B, Shimosato Y, Brambilla E. WHO International Histological Classification of Tumors: Histological Typing of Lung and Pleural Tumors. Heidelberg: Springer; 1999.
-
- Colby T V, Koss M N, Travis W D. Atlas of Tumor Pathology (3rd Series Fascicle 13): Tumors of the Lower Respiratory Tract. under the auspices of Universities Associated for Research and Education in Pathology, Washington, DC: Armed Forces Institute of Pathology; 1995.
-
- Alizadeh A A, Eisen M B, Davis R E, Ma C, Lossos I S, Rosenwald A, Boldrick J C, Sabet H, Tran T, Yu X, et al. Nature (London) 2000;403:503–511. - PubMed
-
- Perou C M, Sorlie T, Eisen M B, van de Rijn M, Jeffrey S S, Rees C A, Pollack J R, Ross D T, Johnsen H, Akslen L A, et al. Nature (London) 2000;406:747–752. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

