Gene yerP, involved in surfactin self-resistance in Bacillus subtilis
- PMID: 11709341
- PMCID: PMC90870
- DOI: 10.1128/AAC.45.12.3566-3573.2001
Gene yerP, involved in surfactin self-resistance in Bacillus subtilis
Abstract
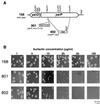
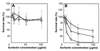
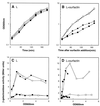
Surfactin is a cyclic lipopeptide biosurfactant. Transposon mutagenesis was performed in Bacillus subtilis strain 168, and a surfactin-susceptible mutant, strain 801, was isolated. Analysis of the region of insertion revealed that yerP was the determinant of surfactin self-resistance. YerP had homology with the resistance, nodulation, and cell division (RND) family proton motive force-dependent efflux pumps only characterized in gram-negative strains. The yerP-deficient strain 802, in which the internal region of the yerP gene of B. subtilis strain 168 was deleted, showed susceptibility to acriflavine and ethidium bromide. When strain 802 was converted to a surfactin producer by introducing a functional sfp which encodes a 4'-phosphopantetheinyl transferase and is mutated in B. subtilis strain 168, this yerP-deficient strain produced surfactin, although surfactin production was significantly reduced. The expression of yerP was at its maximum at the end of the logarithmic growth phase and was not induced by surfactin. yerP is the first RND-like gene characterized in gram-positive strains and is supposed to be involved in the efflux of surfactin.
Figures


References
-
- Arima K, Kakinuma A, Tamura G. Surfactin, a crystalline peptide-lipid surfactant produced by Bacillus subtilis: isolation, characterization and its inhibition of fibrin clot formation. Biochim Biophys Res Commun. 1968;31:488–494. - PubMed
-
- Baev N, Endre G, Petrovics G, Banfalvi Z, Kondorosi A. Six nodulation genes of nod box locus 4 in Rhizobium meliloti are involved in nodulation signal production: nodM codes for D-glucosamine synthetase. Mol Gen Genet. 1991;228:113–124. - PubMed
-
- Beven L, Wroblewski H. Effect of natural amphipathic peptides on viability, membrane potential, cell shape and motility of mollicutes. Res Microbiol. 1997;148:163–175. - PubMed
-
- Cosmina P, Rodriguez F, de Ferra F, Grandi G, Perego M, Venema G, van Sinderen D. Sequence and analysis of the genetic locus responsible for surfactin synthesis in Bacillus subtilis. Mol Microbiol. 1993;8:821–831. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

