Characterization of novel simian immunodeficiency viruses from red-capped mangabeys from Nigeria (SIVrcmNG409 and -NG411)
- PMID: 11711592
- PMCID: PMC116097
- DOI: 10.1128/JVI.75.24.12014-12027.2001
Characterization of novel simian immunodeficiency viruses from red-capped mangabeys from Nigeria (SIVrcmNG409 and -NG411)
Abstract
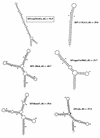
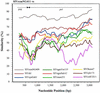
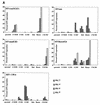
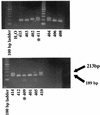
Two novel simian immunodeficiency virus (SIV) strains from wild-caught red-capped mangabeys (Cercocebus torquatus torquatus) from Nigeria were characterized. Sequence analysis of the fully sequenced SIV strain rcmNG411 (SIVrcmNG411) and gag and pol sequence of SIVrcmNG409 revealed that they were genetically most closely related to the recently characterized SIVrcm from Gabon (SIVrcmGB1). Thus, red-capped mangabeys from distant geographic locations harbor a common lineage of SIV. SIVrcmNG411 carried a vpx gene in addition to vpr, suggesting a common evolutionary ancestor with SIVsm (from sooty mangabeys). However, SIVrcm was only marginally closer to SIVsm in that region than to any of the other lentiviruses. SIVrcm showed the highest similarity in pol with SIVdrl, isolated from a drill, a primate that is phylogenetically distinct from mangabey monkeys, and clustered with other primate lentiviruses (primarily SIVcpz [from chimpanzees] and SIVagmSab [from African green monkeys]) discordantly in different regions of the genome, suggesting a history of recombination. Despite the genetic relationship to SIVcpz in the pol gene, SIVrcmNG411 did not replicate in chimpanzee peripheral blood mononuclear cells (PBMC), although two other viruses unrelated to SIVcpz, SIVmndGB1 (from mandrills) and SIVlhoest (from L'Hoest monkeys), were able to grow in chimpanzee PBMC. The CCR5 24-bp deletion previously described in red-capped mangabeys from Gabon was also observed in Nigerian red-capped mangabeys, and SIVrcmNG411, like SIVrcmGB1, used CCR2B and STRL33 as coreceptors for virus entry. SIVrcm, SIVsm, SIVmndGB1, and all four SIVlhoest isolates but not SIVsun (from sun-tailed monkeys) replicated efficiently in human PBMC, suggesting that the ability to infect the human host can vary within one lineage.
Figures





References
-
- Baier M, Werner A, Cichutek K, Garber C, Mueller C, Kraus G, Ferdinand F J, Hartung S, Papas T S, Kurth R. Molecularly cloned simian immunodeficiency virus SIVagm3 is highly divergent from other SIVagm isolates and is biologically active in vitro and in vivo. J Virol. 1989;63:5119–5123. - PMC - PubMed
-
- Beer B E, Bailes E, Dapolito G, Campbell B J, Goeken R M, Axthelm M K, Markham P D, Bernard J, Zagury D, Franchini G, Sharp P M, Hirsch V M. Patterns of genomic sequence diversity among their simian immunodeficiency viruses suggest that L'Hoest monkeys (Cercopithecus lhoesti) are a natural lentivirus reservoir. J Virol. 2000;74:3892–3898. - PMC - PubMed
-
- Beer B E, Bailes E, Goeken R, Dapolito G, Coulibaly C, Norley S G, Kurth R, Gautier J P, Gautier-Hion A, Vallet D, Sharp P M, Hirsch V M. Simian immunodeficiency virus (SIV) from sun-tailed monkeys (Cercopithecus solatus): evidence for host-dependent evolution of SIV within the C. lhoesti superspecies. J Virol. 1999;73:7734–7744. - PMC - PubMed
-
- Beer B E, Bailes E, Sharp P M, Hirsch V M. Diversity and evolution of primate lentiviruses. In: Kuiken C L, Foley B, Hahn B, Korber B, McCutchan F, Marx P A, Mellors J W, Mullins J I, Sodroski J, Wolinksy S, editors. Human retroviruses and AIDS 1999: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.Mex: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; 1999.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

