Sequences in the 5' nontranslated region of hepatitis C virus required for RNA replication
- PMID: 11711595
- PMCID: PMC116100
- DOI: 10.1128/JVI.75.24.12047-12057.2001
Sequences in the 5' nontranslated region of hepatitis C virus required for RNA replication
Abstract
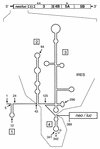
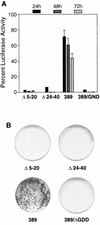
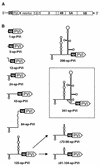
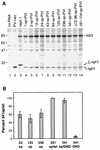
Sequences in the 5' and 3' termini of plus-strand RNA viruses harbor cis-acting elements important for efficient translation and replication. In case of the hepatitis C virus (HCV), a plus-strand RNA virus of the family Flaviviridae, a 341-nucleotide-long nontranslated region (NTR) is located at the 5' end of the genome. This sequence contains an internal ribosome entry site (IRES) that is located downstream of an about 40-nucleotide-long sequence of unknown function. By using our recently developed HCV replicon system, we mapped and characterized the sequences in the 5' NTR required for RNA replication. We show that deletions introduced into the 5' terminal 40 nucleotides abolished RNA replication but only moderately affected translation. By generating a series of replicons with HCV-poliovirus (PV) chimeric 5' NTRs, we could show that the first 125 nucleotides of the HCV genome are essential and sufficient for RNA replication. However, the efficiency could be tremendously increased upon the addition of the complete HCV 5' NTR. These data show that (i) sequences upstream of the HCV IRES are essential for RNA replication, (ii) the first 125 nucleotides of the HCV 5' NTR are sufficient for RNA replication, but such replicon molecules are severely impaired for multiplication, and (iii) high-level HCV replication requires sequences located within the IRES. These data provide the first identification of signals in the 5' NTR of HCV RNA essential for replication of this virus.
Figures

Similar articles
-
Domains I and II in the 5' nontranslated region of the HCV genome are required for RNA replication.Biochem Biophys Res Commun. 2002 Jan 11;290(1):105-12. doi: 10.1006/bbrc.2001.6167. Biochem Biophys Res Commun. 2002. PMID: 11779140
-
Hairpin ribozymes in combination with siRNAs against highly conserved hepatitis C virus sequence inhibit RNA replication and protein translation from hepatitis C virus subgenomic replicons.FEBS J. 2005 Nov;272(22):5910-22. doi: 10.1111/j.1742-4658.2005.04986.x. FEBS J. 2005. PMID: 16279954
-
Isolation of RNA aptamers specific for the HCV minus-IRES domain I.Nucleic Acids Symp Ser (Oxf). 2007;(51):393-4. doi: 10.1093/nass/nrm197. Nucleic Acids Symp Ser (Oxf). 2007. PMID: 18029752
-
[Structure and function of the non-coding regions of hepatitis C viral RNA].Postepy Biochem. 2006;52(1):62-71. Postepy Biochem. 2006. PMID: 16869303 Review. Polish.
-
Translation of hepatitis C virus genome.Princess Takamatsu Symp. 1995;25:99-110. Princess Takamatsu Symp. 1995. PMID: 8875614 Review.
Cited by
-
Base pairing between hepatitis C virus RNA and microRNA 122 3' of its seed sequence is essential for genome stabilization and production of infectious virus.J Virol. 2012 Jul;86(13):7372-83. doi: 10.1128/JVI.00513-12. Epub 2012 Apr 24. J Virol. 2012. PMID: 22532678 Free PMC article.
-
Ultrastructural and biophysical characterization of hepatitis C virus particles produced in cell culture.J Virol. 2010 Nov;84(21):10999-1009. doi: 10.1128/JVI.00526-10. Epub 2010 Aug 4. J Virol. 2010. PMID: 20686033 Free PMC article.
-
Identification of a structural element of the hepatitis C virus minus strand RNA involved in the initiation of RNA synthesis.Nucleic Acids Res. 2010 Jul;38(12):4079-91. doi: 10.1093/nar/gkq109. Epub 2010 Mar 1. Nucleic Acids Res. 2010. PMID: 20194114 Free PMC article.
-
Molecular biology of hepatitis C virus.J Gastroenterol. 2007 Jun;42(6):411-23. doi: 10.1007/s00535-007-2030-3. Epub 2007 Jun 29. J Gastroenterol. 2007. PMID: 17671755 Review.
-
Poly(C)-binding protein 2 interacts with sequences required for viral replication in the hepatitis C virus (HCV) 5' untranslated region and directs HCV RNA replication through circularizing the viral genome.J Virol. 2011 Aug;85(16):7954-64. doi: 10.1128/JVI.00339-11. Epub 2011 Jun 1. J Virol. 2011. PMID: 21632751 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

