The major human immunodeficiency virus type 2 (HIV-2) packaging signal is present on all HIV-2 RNA species: cotranslational RNA encapsidation and limitation of Gag protein confer specificity
- PMID: 11711596
- PMCID: PMC116101
- DOI: 10.1128/JVI.75.24.12058-12069.2001
The major human immunodeficiency virus type 2 (HIV-2) packaging signal is present on all HIV-2 RNA species: cotranslational RNA encapsidation and limitation of Gag protein confer specificity
Abstract
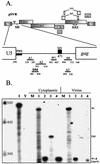
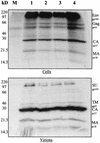
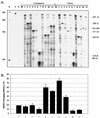
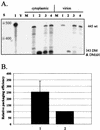
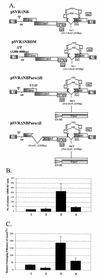
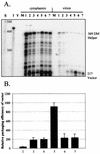
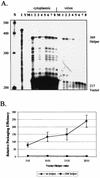
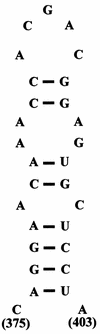
Deletion of a region of the human immunodeficiency virus type 2 (HIV-2) 5' leader RNA reduces genomic RNA encapsidation to about 5% that of wild-type virus with no defect in viral protein production but severely limits virus spread in Jurkat T cells, indicating that this region contains a major cis-acting encapsidation signal, or psi (Psi). Being upstream of the major splice donor, it is present on all viral transcripts. We have shown that HIV-2 selects its genomic RNA for encapsidation cotranslationally, rendering wild-type HIV-2 unable to encapsidate vector RNAs in trans. Virus with Psi deleted, however, encapsidates an HIV-2 vector, demonstrating competition for Gag protein. HIV-2 overcomes the lack of packaging signal location specificity by two novel mechanisms, cotranslational packaging and competition for limiting Gag polyprotein.
Figures


Similar articles
-
Nonreciprocal packaging of human immunodeficiency virus type 1 and type 2 RNA: a possible role for the p2 domain of Gag in RNA encapsidation.J Virol. 1998 Jul;72(7):5877-85. doi: 10.1128/JVI.72.7.5877-5885.1998. J Virol. 1998. PMID: 9621049 Free PMC article.
-
Dimerisation of HIV-2 genomic RNA is linked to efficient RNA packaging, normal particle maturation and viral infectivity.Retrovirology. 2007 Dec 13;4:90. doi: 10.1186/1742-4690-4-90. Retrovirology. 2007. PMID: 18078509 Free PMC article.
-
Human immunodeficiency virus type 2 (HIV-2): packaging signal and associated negative regulatory element.Hum Gene Ther. 1995 Feb;6(2):177-84. doi: 10.1089/hum.1995.6.2-177. Hum Gene Ther. 1995. PMID: 7734518
-
HIV-1 RNA genome packaging: it's G-rated.mBio. 2024 Apr 10;15(4):e0086123. doi: 10.1128/mbio.00861-23. Epub 2024 Feb 27. mBio. 2024. PMID: 38411060 Free PMC article. Review.
-
On the Selective Packaging of Genomic RNA by HIV-1.Viruses. 2016 Sep 12;8(9):246. doi: 10.3390/v8090246. Viruses. 2016. PMID: 27626441 Free PMC article. Review.
Cited by
-
Human immunodeficiency virus type 2 Gag interacts specifically with PRP4, a serine-threonine kinase, and inhibits phosphorylation of splicing factor SF2.J Virol. 2004 Oct;78(20):11303-12. doi: 10.1128/JVI.78.20.11303-11312.2004. J Virol. 2004. PMID: 15452250 Free PMC article.
-
Impaired RNA incorporation and dimerization in live attenuated leader-variants of SIVmac239.Retrovirology. 2006 Dec 21;3:96. doi: 10.1186/1742-4690-3-96. Retrovirology. 2006. PMID: 17184529 Free PMC article.
-
HIV-2 RNA dimerization is regulated by intramolecular interactions in vitro.RNA. 2007 Aug;13(8):1341-54. doi: 10.1261/rna.483807. Epub 2007 Jun 25. RNA. 2007. PMID: 17592043 Free PMC article.
-
Encapsidation determinants located downstream of the major splice donor in the maedi-visna virus leader region.J Virol. 2006 Dec;80(23):11743-55. doi: 10.1128/JVI.01284-06. Epub 2006 Sep 13. J Virol. 2006. PMID: 16971429 Free PMC article.
-
High frequency of genetic recombination is a common feature of primate lentivirus replication.J Virol. 2006 Oct;80(19):9651-8. doi: 10.1128/JVI.00936-06. J Virol. 2006. PMID: 16973569 Free PMC article.
References
-
- Arya S K, Zamani M, Kundra P. Human immunodeficiency virus type 2 lentivirus vectors for gene transfer: expression and potential for helper virus-free packaging. Hum Gene Ther. 1998;9:1371–1380. - PubMed
-
- Berkowitz R D, Hammarskjold M L, Helga-Maria C, Rekosh D, Goff S P. 5′ regions of HIV-1 RNAs are not sufficient for encapsidation: implications for the HIV-1 packaging signal. Virology. 1995;212:718–723. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

