Secondary structural elements within the 3' untranslated region of mouse hepatitis virus strain JHM genomic RNA
- PMID: 11711601
- PMCID: PMC116106
- DOI: 10.1128/JVI.75.24.12105-12113.2001
Secondary structural elements within the 3' untranslated region of mouse hepatitis virus strain JHM genomic RNA
Abstract
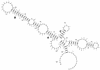
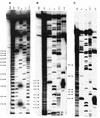
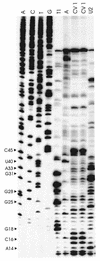
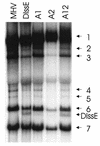
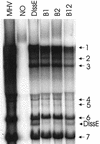
Previously, we characterized two host protein binding elements located within the 3'-terminal 166 nucleotides of the mouse hepatitis virus (MHV) genome and assessed their functions in defective-interfering (DI) RNA replication. To determine the role of RNA secondary structures within these two host protein binding elements in viral replication, we explored the secondary structure of the 3'-terminal 166 nucleotides of the MHV strain JHM genome using limited RNase digestion assays. Our data indicate that multiple stem-loop and hairpin-loop structures exist within this region. Mutant and wild-type DIssEs were employed to test the function of secondary structure elements in DI RNA replication. Three stem structures were chosen as targets for the introduction of transversion mutations designed to destroy base pairing structures. Mutations predicted to destroy the base pairing of nucleotides 142 to 136 with nucleotides 68 to 74 exhibited a deleterious effect on DIssE replication. Destruction of base pairing between positions 96 to 99 and 116 to 113 also decreased DI RNA replication. Mutations interfering with the pairing of nucleotides 67 to 63 with nucleotides 52 to 56 had only minor effects on DIssE replication. The introduction of second complementary mutations which restored the predicted base pairing of positions 142 to 136 with 68 to 74 and nucleotides 96 to 99 with 116 to 113 largely ameliorated defects in replication ability, restoring DI RNA replication to levels comparable to that of wild-type DIssE RNA, suggesting that these secondary structures are important for efficient MHV replication. We also identified a conserved 23-nucleotide stem-loop structure involving nucleotides 142 to 132 and nucleotides 68 to 79. The upstream side of this conserved stem-loop is contained within a host protein binding element (nucleotides 166 to 129).
Figures
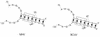
Similar articles
-
A specific host cellular protein binding element near the 3' end of mouse hepatitis virus genomic RNA.Virology. 1997 May 26;232(1):74-85. doi: 10.1006/viro.1997.8553. Virology. 1997. PMID: 9185590
-
Effect of mutations in the mouse hepatitis virus 3'(+)42 protein binding element on RNA replication.J Virol. 2005 Dec;79(23):14570-85. doi: 10.1128/JVI.79.23.14570-14585.2005. J Virol. 2005. PMID: 16282457 Free PMC article.
-
Specific binding of host cellular proteins to multiple sites within the 3' end of mouse hepatitis virus genomic RNA.J Virol. 1995 Apr;69(4):2016-23. doi: 10.1128/JVI.69.4.2016-2023.1995. J Virol. 1995. PMID: 7884846 Free PMC article.
-
[Structure and function of the non-coding regions of hepatitis C viral RNA].Postepy Biochem. 2006;52(1):62-71. Postepy Biochem. 2006. PMID: 16869303 Review. Polish.
-
Functions of the 3' and 5' genome RNA regions of members of the genus Flavivirus.Virus Res. 2015 Aug 3;206:108-19. doi: 10.1016/j.virusres.2015.02.006. Epub 2015 Feb 13. Virus Res. 2015. PMID: 25683510 Free PMC article. Review.
Cited by
-
Functional analysis of the stem loop S3 and S4 structures in the coronavirus 3'UTR.Virology. 2013 Aug 15;443(1):40-7. doi: 10.1016/j.virol.2013.04.021. Epub 2013 May 17. Virology. 2013. PMID: 23683838 Free PMC article.
-
A U-turn motif-containing stem-loop in the coronavirus 5' untranslated region plays a functional role in replication.RNA. 2007 May;13(5):763-80. doi: 10.1261/rna.261807. Epub 2007 Mar 12. RNA. 2007. PMID: 17353353 Free PMC article.
-
Stem-loop III in the 5' untranslated region is a cis-acting element in bovine coronavirus defective interfering RNA replication.J Virol. 2003 Jun;77(12):6720-30. doi: 10.1128/jvi.77.12.6720-6730.2003. J Virol. 2003. PMID: 12767992 Free PMC article.
-
Genetic interactions between an essential 3' cis-acting RNA pseudoknot, replicase gene products, and the extreme 3' end of the mouse coronavirus genome.J Virol. 2008 Feb;82(3):1214-28. doi: 10.1128/JVI.01690-07. Epub 2007 Nov 21. J Virol. 2008. PMID: 18032506 Free PMC article.
-
The 3'-terminal 55 nucleotides of bovine coronavirus defective interfering RNA harbor cis-acting elements required for both negative- and positive-strand RNA synthesis.PLoS One. 2014 May 22;9(5):e98422. doi: 10.1371/journal.pone.0098422. eCollection 2014. PLoS One. 2014. PMID: 24852421 Free PMC article.
References
-
- Agrawal S, Gupta D, Panda S K. The 3′ end of hepatitis E virus (HEV) genome binds specifically to the viral RNA-dependent RNA polymerase (RdRp) Virology. 2001;282:87–101. - PubMed
-
- Barthold S W. Host age and genotypic effects on enterotropic mouse hepatitis virus infection. Lab Anim Sci. 1987;37:36–40. - PubMed
-
- Chen D, Barros M, Spencer E, Patton J T. Features of the 3′-consensus sequence of rotavirus mRNAs critical to minus strand synthesis. Virology. 2001;282:221–229. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

