Ribin, a protein encoded by a message complementary to rRNA, modulates ribosomal transcription and cell proliferation
- PMID: 11713263
- PMCID: PMC99991
- DOI: 10.1128/MCB.21.24.8255-8263.2001
Ribin, a protein encoded by a message complementary to rRNA, modulates ribosomal transcription and cell proliferation
Abstract
The control of rRNA transcription, tightly coupled to the cell cycle and growth state of the cell, is a key process for understanding the mechanisms that drive cell proliferation. Here we describe a novel protein, ribin, found in rodents, that binds to the rRNA promoter and stimulates its activity. The protein also interacts with the basal rRNA transcription factor UBF. The open reading frame encoding ribin is 96% complementary to a central region of the large rRNA. This demonstrates that ribosomal DNA-related sequences in higher eukaryotes can be expressed as protein-coding messages. Ribin contains two predicted nuclear localization sequence elements, and green fluorescent protein-ribin fusion proteins localize in the nucleus. Cell lines overexpressing ribin exhibit enhanced rRNA transcription and faster growth. Furthermore, these cells significantly overcome the suppression of rRNA synthesis caused by serum deprivation. On the other hand, the endogenous ribin level correlates positively with the amount of serum in the medium. The data show that ribin is a limiting stimulatory factor for rRNA synthesis in vivo and suggest its involvement in the pathway that adapts ribosomal transcription and cell proliferation to physiological changes.
Figures


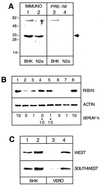


Similar articles
-
Transcription factor BACH1 is recruited to the nucleus by its novel alternative spliced isoform.J Biol Chem. 2001 Mar 9;276(10):7278-84. doi: 10.1074/jbc.M004227200. Epub 2000 Nov 7. J Biol Chem. 2001. PMID: 11069897
-
Nucleomorphin. A novel, acidic, nuclear calmodulin-binding protein from dictyostelium that regulates nuclear number.J Biol Chem. 2002 May 31;277(22):19735-44. doi: 10.1074/jbc.M109717200. Epub 2002 Mar 27. J Biol Chem. 2002. PMID: 11919178
-
Nog2p, a putative GTPase associated with pre-60S subunits and required for late 60S maturation steps.EMBO J. 2001 Nov 15;20(22):6475-84. doi: 10.1093/emboj/20.22.6475. EMBO J. 2001. PMID: 11707418 Free PMC article.
-
SV40 large T antigen binds to the TBP-TAF(I) complex SL1 and coactivates ribosomal RNA transcription.Genes Dev. 1997 Jun 15;11(12):1605-17. doi: 10.1101/gad.11.12.1605. Genes Dev. 1997. PMID: 9203586
-
[rRNA transcription and metabolic stress response].Seikagaku. 2009 Jun;81(6):456-64. Seikagaku. 2009. PMID: 19618869 Review. Japanese. No abstract available.
Cited by
-
Misannotations of rRNA can now generate 90% false positive protein matches in metatranscriptomic studies.Nucleic Acids Res. 2011 Nov 1;39(20):8792-802. doi: 10.1093/nar/gkr576. Epub 2011 Jul 19. Nucleic Acids Res. 2011. PMID: 21771858 Free PMC article.
-
Small RNAs from mitochondrial genome recombination sites are incorporated into T. gondii mitoribosomes.Elife. 2024 Feb 16;13:e95407. doi: 10.7554/eLife.95407. Elife. 2024. PMID: 38363119 Free PMC article.
-
The Ribosome as a Missing Link in Prebiotic Evolution III: Over-Representation of tRNA- and rRNA-Like Sequences and Plieofunctionality of Ribosome-Related Molecules Argues for the Evolution of Primitive Genomes from Ribosomal RNA Modules.Int J Mol Sci. 2019 Jan 2;20(1):140. doi: 10.3390/ijms20010140. Int J Mol Sci. 2019. PMID: 30609737 Free PMC article.
-
A novel mitochondrial protein, Tar1p, is encoded on the antisense strand of the nuclear 25S rDNA.Genes Dev. 2002 Nov 1;16(21):2755-60. doi: 10.1101/gad.1035002. Genes Dev. 2002. PMID: 12414727 Free PMC article.
-
Regulating ehrlich and demethiolation pathways for alcohols production by the expression of ubiquitin-protein ligase gene HUWE1.Sci Rep. 2016 Feb 10;6:20828. doi: 10.1038/srep20828. Sci Rep. 2016. PMID: 26860895 Free PMC article.
References
-
- Abath F, Simpson A. A simple method for the recovery of purified recombinant peptides cleaved from glutathione-S-transferase fusion proteins. Peptide Res. 1990;3:167–168. - PubMed
-
- Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1992.
-
- Bachellerie J-P, Michot B, Nicoloso M, Balakin A, Ni J, Fournier M. Antisense snoRNAs: a family of nucleolar RNAs with long complementarities to RNA. Trends Biochem Sci. 1995;20:261–264. - PubMed
-
- Beckmann H, Chen J-L, O'Brien T, Tjian R. Coactivator and promoter-selective properties of RNA polymerase I TAFs. Science. 1995;270:1506–1509. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
