Involvement of proteasome alpha-subunit PSMA7 in hepatitis C virus internal ribosome entry site-mediated translation
- PMID: 11713272
- PMCID: PMC100000
- DOI: 10.1128/MCB.21.24.8357-8364.2001
Involvement of proteasome alpha-subunit PSMA7 in hepatitis C virus internal ribosome entry site-mediated translation
Abstract
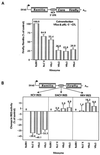
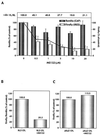
Ribozymes are small catalytic RNA molecules that can be engineered to enzymatically cleave RNA transcripts in a sequence-specific fashion and thereby inhibit expression and function of the corresponding gene product. With their simple structures and site-specific cleavage activity, they have been exploited as potential therapeutic agents in a variety of human disorders, including hepatitis C virus (HCV) infection. We have designed a hairpin ribozyme (Rz3'X) targeting the HCV minus-strand replication intermediate at position 40 within the 3'X tail. Surprisingly, Rz3'X was found to induce ganciclovir (GCV)-resistant colonies in a bicistronic cellular reporter system with HCV internal ribosome entry site (IRES)-dependent translation of herpes simplex virus thymidine kinase (TK). Rz3'X-transduced GCV-resistant HeLa reporter cells showed substantially reduced IRES-mediated HCV core protein translation compared with control vector-transduced cells. Since these reporter systems do not contain the HCV 3'X tail sequences, the results indicate that Rz3'X probably exerted an inhibitory effect on HCV IRES activity fortuitously through another gene target. A novel technique of ribozyme cleavage-based target gene identification (cleavage-specific amplification of cDNA ends) (M. Krüger, C. Beger, P. J. Welch, J. R. Barber, and F. Wong-Staal, Nucleic Acids Res. 29:e94, 2001) revealed that human 20S proteasome alpha-subunit PSMA7 mRNA was a target RNA recognized and cleaved by Rz3'X. We then showed that additional ribozymes directed against PSMA7 RNA inhibited HCV IRES activity in two assay systems: GCV resistance in the HeLa IRES TK reporter cell system and a transient transfection assay performed with a bicistronic Renilla-HCV IRES-firefly luciferase reporter in Huh7 cells. In contrast, ribozymes were inactive against IRES of encephalomyocarditis virus and human rhinovirus. Additionally, proteasome inhibitor MG132 exerted a dose-dependent inhibitory effect on HCV IRES-mediated translation but not on cap-dependent translation. These data suggest a principal role for PSMA7 in regulating HCV IRES activity, a function essential for HCV replication.
Figures




References
-
- Adams J, Palombella V J, Elliott P J. Proteasome inhibition: a new strategy in cancer treatment. Investig New Drugs. 2000;18:109–121. - PubMed
-
- Beger C, Krüger M, Wong-Staal F. Ribozymes in cancer gene therapy. In: Lattime E C, Gerson S L, editors. Gene therapy of cancer. San Diego, Calif: Academic Press; 1998. pp. 139–152.
-
- Blight K J, Kolykhalov A A, Rice C M. Efficient initiation of HCV RNA replication in cell culture. Science. 2000;290:1972–1974. - PubMed
-
- Goldberg A L, Rock K L. Proteolysis, proteasomes and antigen presentation. Nature. 1992;357:375–379. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
