Domain structure of the NRIF3 family of coregulators suggests potential dual roles in transcriptional regulation
- PMID: 11713274
- PMCID: PMC100002
- DOI: 10.1128/MCB.21.24.8371-8384.2001
Domain structure of the NRIF3 family of coregulators suggests potential dual roles in transcriptional regulation
Abstract
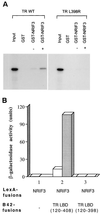
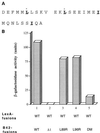
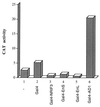
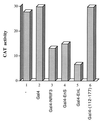
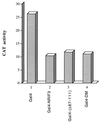
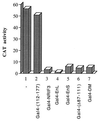
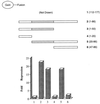
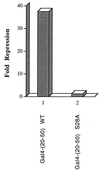
The identification of a novel coregulator for nuclear hormone receptors, designated NRIF3, was recently reported (D. Li et al., Mol. Cell. Biol. 19:7191-7202, 1999). Unlike most known coactivators, NRIF3 exhibits a distinct receptor specificity in interacting with and potentiating the activity of only TRs and RXRs but not other examined nuclear receptors. However, the molecular basis underlying such specificity is unclear. In this report, we extended our study of NRIF3-receptor interactions. Our results suggest a bivalent interaction model, where a single NRIF3 molecule utilizes both the C-terminal LXXIL (receptor-interacting domain 1 [RID1]) and the N-terminal LXXLL (RID2) modules to cooperatively interact with TR or RXR (presumably a receptor dimer), with the spacing between RID1 and RID2 playing an important role in influencing the affinity of the interactions. During the course of these studies, we also uncovered an NRIF3-NRIF3 interaction domain. Deletion and mutagenesis analyses mapped the dimerization domain to a region in the middle of NRIF3 (residues 84 to 112), which is predicted to form a coiled-coil structure and contains a putative leucine zipper-like motif. By using Gal4 fusion constructs, we identified an autonomous transactivation domain (AD1) at the C terminus of NRIF3. Somewhat surprisingly, full-length NRIF3 fused to the DNA-binding domain of Gal4 was found to repress transcription of a Gal4 reporter. Further analyses mapped a novel repression domain (RepD1) to a small region at the N-terminal portion of NRIF3 (residues 20 to 50). The NRIF3 gene encodes at least two additional isoforms due to alternative splicing. These two isoforms contain the same RepD1 region as NRIF3. Consistent with this, Gal4 fusions of these two isoforms were also found to repress transcription. Cotransfection of NRIF3 or its two isoforms did not relieve the transrepression function mediated by their corresponding Gal4 fusion proteins, suggesting that the repression involves a mechanism(s) other than the recruitment of a titratable corepressor. Interestingly, a single amino acid residue change of a potential phosphorylation site in RepD1 (Ser(28) to Ala) abolishes its transrepression function, suggesting that the coregulatory property of NRIF3 (or its isoforms) might be subjected to regulation by cellular signaling. Taken together, our results identify NRIF3 as an interesting coregulator that possesses both transactivation and transrepression domains and/or functions. Collectively, the NRIF3 family of coregulators (which includes NRIF3 and its other isoforms) may play dual roles in mediating both positive and negative regulatory effects on gene expression.
Figures





Similar articles
-
Metastasis-associated protein 1 interacts with NRIF3, an estrogen-inducible nuclear receptor coregulator.Mol Cell Biol. 2004 Aug;24(15):6581-91. doi: 10.1128/MCB.24.15.6581-6591.2004. Mol Cell Biol. 2004. PMID: 15254226 Free PMC article.
-
NRIF3 is a novel coactivator mediating functional specificity of nuclear hormone receptors.Mol Cell Biol. 1999 Oct;19(10):7191-202. doi: 10.1128/MCB.19.10.7191. Mol Cell Biol. 1999. PMID: 10490654 Free PMC article.
-
The ligand-binding domains of the thyroid hormone/retinoid receptor gene subfamily function in vivo to mediate heterodimerization, gene silencing, and transactivation.Mol Cell Biol. 1995 Mar;15(3):1817-25. doi: 10.1128/MCB.15.3.1817. Mol Cell Biol. 1995. PMID: 7862171 Free PMC article.
-
Gene silencing by chicken ovalbumin upstream promoter-transcription factor I (COUP-TFI) is mediated by transcriptional corepressors, nuclear receptor-corepressor (N-CoR) and silencing mediator for retinoic acid receptor and thyroid hormone receptor (SMRT).Mol Endocrinol. 1997 Jun;11(6):714-24. doi: 10.1210/mend.11.6.0002. Mol Endocrinol. 1997. PMID: 9171235
-
The dimerization/repression domain of RFX1 is related to a conserved region of its yeast homologues Crt1 and Sak1: a new function for an ancient motif.J Mol Biol. 1999 Nov 19;294(1):121-37. doi: 10.1006/jmbi.1999.3245. J Mol Biol. 1999. PMID: 10556033
Cited by
-
Regulation of alternative splicing of Gtf2ird1 and its impact on slow muscle promoter activity.Biochem J. 2003 Sep 1;374(Pt 2):359-67. doi: 10.1042/BJ20030189. Biochem J. 2003. PMID: 12780350 Free PMC article.
-
TNIP1 is a corepressor of agonist-bound PPARs.Arch Biochem Biophys. 2011 Dec 1;516(1):58-66. doi: 10.1016/j.abb.2011.08.014. Epub 2011 Sep 22. Arch Biochem Biophys. 2011. PMID: 21967852 Free PMC article.
-
Metastasis-associated protein 1 interacts with NRIF3, an estrogen-inducible nuclear receptor coregulator.Mol Cell Biol. 2004 Aug;24(15):6581-91. doi: 10.1128/MCB.24.15.6581-6591.2004. Mol Cell Biol. 2004. PMID: 15254226 Free PMC article.
-
Long-read isoform sequencing reveals tissue-specific isoform expression between active and hibernating brown bears (Ursus arctos).G3 (Bethesda). 2022 Mar 4;12(3):jkab422. doi: 10.1093/g3journal/jkab422. G3 (Bethesda). 2022. PMID: 35100340 Free PMC article.
-
Identification of a novel pathway that selectively modulates apoptosis of breast cancer cells.Cancer Res. 2009 Feb 15;69(4):1375-82. doi: 10.1158/0008-5472.CAN-08-2896. Epub 2009 Feb 3. Cancer Res. 2009. PMID: 19190336 Free PMC article.
References
-
- Anzick S L, Kononen J, Walker R L, Azorsa D O, Tanner M M, Guan X Y, Sauter G, Kallioniemi O P, Trent J M, Meltzer P S. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. - PubMed
-
- Carson-Jurica M A, Schrader W T, O'Malley B W. Steroid receptor family: structure and functions. Endocrine Rev. 1990;11:201–218. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
