Ribosomal pausing at a frameshifter RNA pseudoknot is sensitive to reading phase but shows little correlation with frameshift efficiency
- PMID: 11713298
- PMCID: PMC100026
- DOI: 10.1128/MCB.21.24.8657-8670.2001
Ribosomal pausing at a frameshifter RNA pseudoknot is sensitive to reading phase but shows little correlation with frameshift efficiency
Abstract
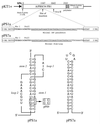
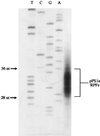
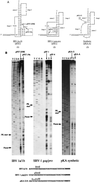
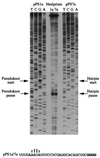
Here we investigated ribosomal pausing at sites of programmed -1 ribosomal frameshifting, using translational elongation and ribosome heelprint assays. The site of pausing at the frameshift signal of infectious bronchitis virus (IBV) was determined and was consistent with an RNA pseudoknot-induced pause that placed the ribosomal P- and A-sites over the slippery sequence. Similarly, pausing at the simian retrovirus 1 gag/pol signal, which contains a different kind of frameshifter pseudoknot, also placed the ribosome over the slippery sequence, supporting a role for pausing in frameshifting. However, a simple correlation between pausing and frameshifting was lacking. Firstly, a stem-loop structure closely related to the IBV pseudoknot, although unable to stimulate efficient frameshifting, paused ribosomes to a similar extent and at the same place on the mRNA as a parental pseudoknot. Secondly, an identical pausing pattern was induced by two pseudoknots differing only by a single loop 2 nucleotide yet with different functionalities in frameshifting. The final observation arose from an assessment of the impact of reading phase on pausing. Given that ribosomes advance in triplet fashion, we tested whether the reading frame in which ribosomes encounter an RNA structure (the reading phase) would influence pausing. We found that the reading phase did influence pausing but unexpectedly, the mRNA with the pseudoknot in the phase which gave the least pausing was found to promote frameshifting more efficiently than the other variants. Overall, these experiments support the view that pausing alone is insufficient to mediate frameshifting and additional events are required. The phase dependence of pausing may be indicative of an activity in the ribosome that requires an optimal contact with mRNA secondary structures for efficient unwinding.
Figures





References
-
- Alam S L, Wills N M, Ingram J A, Atkins J F, Gesteland R F. Structural studies of the RNA pseudoknot required for readthrough of the gag-termination codon of murine leukemia virus. J Mol Biol. 1999;288:837–852. - PubMed
-
- Brierley I. Ribosomal frameshifting on viral RNAs. J Gen Virol. 1995;76:1885–1892. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
