Box C/D RNA guides for the ribose methylation of archaeal tRNAs. The tRNATrp intron guides the formation of two ribose-methylated nucleosides in the mature tRNATrp
- PMID: 11713301
- PMCID: PMC92551
- DOI: 10.1093/nar/29.22.4518
Box C/D RNA guides for the ribose methylation of archaeal tRNAs. The tRNATrp intron guides the formation of two ribose-methylated nucleosides in the mature tRNATrp
Abstract
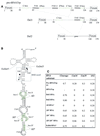
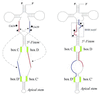
Following a search of the Pyrococcus genomes for homologs of eukaryotic methylation guide small nucleolar RNAs, we have experimentally identified in Pyrococcus abyssi four novel box C/D small RNAs predicted to direct 2'-O-ribose methylations onto the first position of the anticodon in tRNALeu(CAA), tRNALeu(UAA), elongator tRNAMet and tRNATrp, respectively. Remarkably, one of them corresponds to the intron of its presumptive target, pre-tRNATrp. This intron is predicted to direct in cis two distinct ribose methylations within the unspliced tRNA precursor, not only onto the first position of the anticodon in the 5' exon but also onto position 39 (universal tRNA numbering) in the 3' exon. The two intramolecular RNA duplexes expected to direct methylation, which both span an exon-intron junction in pre-tRNATrp, are phylogenetically conserved in euryarchaeotes. We have experimentally confirmed the predicted guide function of the box C/D intron in halophile Haloferax volcanii by mutagenesis analysis, using an in vitro splicing/RNA modification assay in which the two cognate ribose methylations of pre-tRNATrp are faithfully reproduced. Euryarchaeal pre-tRNATrp should provide a unique system to further investigate the molecular mechanisms of RNA-guided ribose methylation and gain new insights into the origin and evolution of the complex family of archaeal and eukaryotic box C/D small RNAs.
Figures




References
-
- Bachellerie J.P. and Cavaille,J. (1997) Guiding ribose methylation of rRNA. Trends Biochem. Sci., 22, 257–261. - PubMed
-
- Smith C.M. and Steitz,J.A. (1997) Sno storm in the nucleolus: new roles for myriad small RNPs. Cell, 89, 669–672. - PubMed
-
- Bachellerie J.P. and Cavaillé,J. (1998) Small nucleolar RNAs guide the ribose methylations of eukaryotic rRNAs. In Grosjean,H. and Benne,R. (eds), Modification and Editing of RNA: The Alteration of RNA Structure and Function. ASM Press, Washington, DC, pp. 255–272.
-
- Bachellerie J.P., Michot,B., Nicoloso,M., Balakin,A., Ni,J. and Fournier,M.J. (1995) Antisense snoRNAs: a family of nucleolar RNAs with long complementarities to rRNA. Trends Biochem. Sci., 20, 261–264. - PubMed
-
- Nicoloso M., Qu,L.H., Michot,B. and Bachellerie,J.P. (1996) Intron-encoded, antisense small nucleolar RNAs: the characterization of nine novel species points to their direct role as guides for the 2′-O-ribose methylation of rRNAs. J. Mol. Biol., 260, 178–195. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions

