Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4): an archaeal DinB-like DNA polymerase with lesion-bypass properties akin to eukaryotic poleta
- PMID: 11713310
- PMCID: PMC92520
- DOI: 10.1093/nar/29.22.4607
Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4): an archaeal DinB-like DNA polymerase with lesion-bypass properties akin to eukaryotic poleta
Abstract
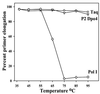
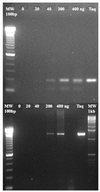
Phylogenetic analysis of Y-family DNA polymerases suggests that it can be subdivided into several discrete branches consisting of UmuC/DinB/Rev1/Rad30/Rad30A and Rad30B. The most diverse is the DinB family that is found in all three kingdoms of life. Searches of the complete genome of the crenarchaeon Sulfolobus solfataricus P2 reveal that it possesses a DinB homolog that has been termed DNA polymerase IV (Dpo4). We have overproduced and purified native Dpo4 protein and report here its enzymatic characterization. Dpo4 is thermostable, but can also synthesize DNA at 37 degrees C. Under these conditions, the enzyme exhibits misinsertion fidelities in the range of 8 x 10(-3) to 3 x 10(-4). Dpo4 is distributive but at high enzyme to template ratios can synthesize long stretches of DNA and can substitute for Taq polymerase in PCR. On damaged DNA templates, Dpo4 can facilitate translesion replication of an abasic site, a cis-syn thymine-thymine dimer, as well as acetyl aminofluorene adducted- and cisplatinated-guanine residues. Thus, although phylogenetically related to DinB polymerases, our studies suggest that the archaeal Dpo4 enzyme exhibits lesion-bypass properties that are, in fact, more akin to those of eukaryotic poleta.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

