A downstream regulatory element located within the coding sequence mediates autoregulated expression of the yeast fatty acid synthase gene FAS2 by the FAS1 gene product
- PMID: 11713312
- PMCID: PMC92567
- DOI: 10.1093/nar/29.22.4625
A downstream regulatory element located within the coding sequence mediates autoregulated expression of the yeast fatty acid synthase gene FAS2 by the FAS1 gene product
Abstract
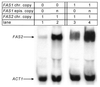
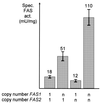
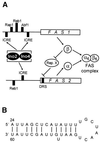
The fatty acid synthase genes FAS1 and FAS2 of the yeast Saccharomyces cerevisiae are transcriptionally co-regulated by general transcription factors (such as Reb1, Rap1 and Abf1) and by the phospholipid-specific heterodimeric activator Ino2/Ino4, acting via their corresponding upstream binding sites. Here we provide evidence for a positive autoregulatory influence of FAS1 on FAS2 expression. Even with a constant FAS2 copy number, a 10-fold increase of FAS2 transcript amount was observed in the presence of FAS1 in multi-copy, compared to a fas1 null mutant. Surprisingly, the first 66 nt of the FAS2 coding region turned out as necessary and sufficient for FAS1-dependent gene expression. FAS2-lacZ fusion constructs deleted for this region showed high reporter gene expression even in the absence of FAS1, arguing for a negatively-acting downstream repression site (DRS) responsible for FAS1-dependent expression of FAS2. Our data suggest that the FAS1 gene product, in addition to its catalytic function, is also required for the coordinate biosynthetic control of the yeast FAS complex. An excess of uncomplexed Fas1 may be responsible for the deactivation of an FAS2-specific repressor, acting via the DRS.
Figures


Similar articles
-
Regulatory gene INO4 of yeast phospholipid biosynthesis is positively autoregulated and functions as a transactivator of fatty acid synthase genes FAS1 and FAS2 from Saccharomyces cerevisiae.Nucleic Acids Res. 1992 Nov 25;20(22):5955-61. doi: 10.1093/nar/20.22.5955. Nucleic Acids Res. 1992. PMID: 1461729 Free PMC article.
-
Importance of general regulatory factors Rap1p, Abf1p and Reb1p for the activation of yeast fatty acid synthase genes FAS1 and FAS2.Eur J Biochem. 1994 Oct 1;225(1):213-22. doi: 10.1111/j.1432-1033.1994.00213.x. Eur J Biochem. 1994. PMID: 7925441
-
Molecular cloning of the yeast fatty acid synthetase genes, FAS1 and FAS2: illustrating the structure of the FAS1 cluster gene by transcript mapping and transformation studies.Mol Gen Genet. 1984;194(3):457-65. doi: 10.1007/BF00425558. Mol Gen Genet. 1984. PMID: 6330502
-
Microbial type I fatty acid synthases (FAS): major players in a network of cellular FAS systems.Microbiol Mol Biol Rev. 2004 Sep;68(3):501-17, table of contents. doi: 10.1128/MMBR.68.3.501-517.2004. Microbiol Mol Biol Rev. 2004. PMID: 15353567 Free PMC article. Review.
-
FIRE (fatty acid synthase insulin-responsive element) and ICE (inverted CCAAT element) regulate fatty acid synthase.Biochem Soc Trans. 1997 Nov;25(4):1220-4. doi: 10.1042/bst0251220. Biochem Soc Trans. 1997. PMID: 9449979 Review. No abstract available.
Cited by
-
Systematic Engineering of Saccharomyces cerevisiae for the De Novo Biosynthesis of Genistein and Glycosylation Derivatives.J Fungi (Basel). 2024 Feb 26;10(3):176. doi: 10.3390/jof10030176. J Fungi (Basel). 2024. PMID: 38535185 Free PMC article.
-
De novo biosynthesis of bioactive isoflavonoids by engineered yeast cell factories.Nat Commun. 2021 Oct 19;12(1):6085. doi: 10.1038/s41467-021-26361-1. Nat Commun. 2021. PMID: 34667183 Free PMC article.
-
QTL mapping of volatile compound production in Saccharomyces cerevisiae during alcoholic fermentation.BMC Genomics. 2018 Mar 1;19(1):166. doi: 10.1186/s12864-018-4562-8. BMC Genomics. 2018. PMID: 29490607 Free PMC article.
-
Transcriptional regulation of phospholipid biosynthesis is linked to fatty acid metabolism by an acyl-CoA-binding-protein-dependent mechanism in Saccharomyces cerevisiae.Biochem J. 2007 Oct 15;407(2):219-30. doi: 10.1042/BJ20070315. Biochem J. 2007. PMID: 17593018 Free PMC article.
-
Universal function-specificity of codon usage.Nucleic Acids Res. 2009 Nov;37(21):7014-23. doi: 10.1093/nar/gkp792. Nucleic Acids Res. 2009. PMID: 19773421 Free PMC article.
References
-
- Struhl K. (1989) Molecular mechanisms of transcriptional regulation in yeast. Annu. Rev. Biochem., 58, 1051–1077. - PubMed
-
- Schweizer M., Roberts,L.M., Höltke,H.-J., Takabayashi,K., Höllerer,E., Hoffmann,B., Müller,G., Köttig,H. and Schweizer,E. (1986) The pentafunctional FAS1 gene of yeast: its nucleotide sequence and order of the catalytic domains. Mol. Gen. Genet., 203, 479–486. - PubMed
-
- Schweizer E., Müller,G., Roberts,L.M., Schweizer,M., Rösch,J., Wiesner,P., Beck,J., Stratmann,D. and Zauner,I. (1987) Genetic control of fatty acid synthetase biosynthesis and structure in lower fungi. Fat Sci. Technol., 89, 570–577.
-
- Chirala S.S., Kuziora,M.A., Spector,D.M. and Wakil,S.J. (1987) Complementation of mutations and nucleotide sequence of FAS1 gene encoding β subunit of yeast fatty acid synthase. J. Biol. Chem., 262, 4231–4240. - PubMed
-
- Mohamed A.H., Chirala,S.S., Mody,N.H., Huang,W.-Y. and Wakil,S.J. (1988) Primary structure of the multifunctional α subunit protein of yeast fatty acid synthase derived from FAS2 gene sequence. J. Biol. Chem., 263, 12315–12325. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

