Quantitative-trait loci influencing body-mass index reside on chromosomes 7 and 13: the National Heart, Lung, and Blood Institute Family Heart Study
- PMID: 11713718
- PMCID: PMC384905
- DOI: 10.1086/338144
Quantitative-trait loci influencing body-mass index reside on chromosomes 7 and 13: the National Heart, Lung, and Blood Institute Family Heart Study
Abstract
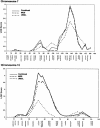
Obesity is a risk factor for many chronic diseases, including glucose intolerance, lipid disorders, hypertension, and coronary heart disease. Even though the body-mass index (BMI) is a heterogeneous phenotype reflecting the amount of fat, lean mass, and body build, several studies have provided evidence of one or two major loci contributing to the variation in this complex trait. We sought to identify loci with potential influence on BMI in the data obtained from National Heart, Lung, and Blood Institute Family Heart Study. Two complementary samples were studied: (a) 1,184 subjects in 317 sibships, with 243 markers typed by the Utah Molecular Genetics Laboratory (UMGL) and (b) 3,027 subjects distributed among 401 three-generation families, with 404 markers typed by the Mammalian Genotyping Service (MGS). A genome scan using a variance-components-based linkage approach was performed for each sample, as well as for the combined sample, in which the markers from each analysis were placed on a common genetic map. There was strong evidence for linkage on chromosome 7q32.3 in each sample: the maximum multipoint LOD scores were 4.7 (P<10-5) at marker GATA43C11 and 3.2 (P=.00007) at marker D7S1804, for the MGS and UMGL samples, respectively. The linkage result is replicated by the consistent evidence from these two complementary subsets. Furthermore, the evidence for linkage was maintained in the combined sample, with a LOD score of 4.9 (P<10-5) for both markers, which map to the same location. This signal is very near the published location for the leptin gene, which is the most prominent candidate gene in this region. For the combined-sample analysis, evidence of linkage was also found on chromosome 13q14, with D13S257 (LOD score 3.2, P=.00006), and other, weaker signals (LOD scores 1.5-1.9) were found on chromosomes 1, 2, 3, 5, 6, 14, and 15.
Figures

References
Electronic-Database Information
-
- ASPEX ftp Site, ftp://lahmed.stanford.edu/pub/aspex/index.html
-
- Center for Medical Genetics, Marshfield Medical Research Foundation, http://research.marshfieldclinic.org/genetics/
-
- Family Heart Study, http://www5.biostat.wustl.edu/fhs/
-
- Genetic Location Database, The, http://cedar.genetics.soton.ac.uk/public_html/ldb.html
-
- Human Genome Project Working Draft at UCSC, http://genome.ucsc.edu/
References
-
- Arita Y, Kihara S, Ouchi N, Takahashi M, Maeda K, Miyagawa J, Hotta K, Shimomura I, Nakamura T, Miyaoka K, Kuriyama H, Nishida M, Yamashita S, Okubo K, Matsubara K, Muraguchi M, Ohmoto Y, Funahashi T, Matsuzawa Y (1999) Paradoxical decrease of an adipose-specific protein, adiponectin, in obesity. Biochem Biophys Res Commun 257:79–83 - PubMed
-
- Atwood LD, Samollow PB, Hixson JE, Stern MP, MacCluer JW (2001) Genome-wide linkage analysis of blood pressure in Mexican Americans. Genet Epidemiol 20:373–382 - PubMed
-
- Aubert R, Betoulle D, Herbeth B, Siest G, Fumeron F (2000) 5-HT2A receptor gene polymorphism is associated with food and alcohol intake in obese people. Int J Obes Relat Metab Disord 24:920–924 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

