Intercellular transfer and supramolecular organization of human leukocyte antigen C at inhibitory natural killer cell immune synapses
- PMID: 11714757
- PMCID: PMC2193674
- DOI: 10.1084/jem.194.10.1507
Intercellular transfer and supramolecular organization of human leukocyte antigen C at inhibitory natural killer cell immune synapses
Erratum in
- J Exp Med 2001 Dec 17;194(12):1883
Abstract
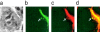
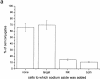
After accumulation of target cell human leukocyte antigen (HLA)-C at inhibitory natural killer (NK) cell immune synapses, some HLA-C transfers from target cells to NK cell plasma membranes and cytoplasm. This unexpected intercellular transfer of HLA-C is dependent on NK receptor recognition, since HLA-Cw6 or -Cw4 but not -Cw3 transfer to an NK transfectant expressing killer Ig-like receptor (KIR)2DL1. Strikingly, live-cell time-lapse laser scanning confocal microscopy shows vesicles containing target cell green fluorescent protein-tagged HLA-C migrating away from immune synapses into NK cells. Unlike clustering of HLA-C at the immune synapse, intercellular transfer of HLA-C is dependent on NK cell ATP, but not target cell ATP. However, the intercellular transfer of HLA-C is not dependent on active polymerization of the actin cytoskeleton. In addition, different arrangements of HLA-C are seen at inhibitory NK immune synapses, and these alter as NK synapses mature, but in a fashion distinct from that seen upon T cell activation.
Figures








References
-
- Ljunggren, H.G., and K. Karre. 1990. In search of the ‘missing self’: MHC molecules and NK cell recognition. Immunol. Today. 11:237–244. - PubMed
-
- Lanier, L.L. 1998. NK cell receptors. Annu. Rev. Immunol. 16:359–393. - PubMed
-
- Long, E.O. 1999. Regulation of immune responses through inhibitory receptors. Annu. Rev. Immunol. 17:875–904. - PubMed
-
- Long, E.O., and S. Rajagopalan. 2000. HLA class I recognition by killer cell Ig-like receptors. Semin. Immunol. 12:101–108. - PubMed

