Structure determination of human and murine beta-defensins reveals structural conservation in the absence of significant sequence similarity
- PMID: 11714914
- PMCID: PMC2374044
- DOI: 10.1110/ps.24401
Structure determination of human and murine beta-defensins reveals structural conservation in the absence of significant sequence similarity
Abstract
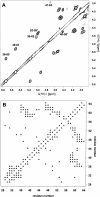
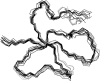
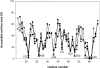
Defensins are cationic and cysteine-rich peptides that play a crucial role in the host defense against microorganisms of many organisms by their capability to permeabilize bacterial membranes. The low sequence similarity among the members of the large mammalian beta-defensin family suggests that their antimicrobial activity is largely independent of their primary structure. To investigate to what extent these defensins share a similar fold, the structures of the two human beta-defensins, hBD-1 and hBD-2, as well as those of two novel murine defensins, termed mBD-7 and mBD-8, were determined by nuclear magnetic resonance spectroscopy. All four defensins investigated share a striking similarity on the level of secondary and tertiary structure including the lack of a distinct hydrophobic core, suggesting that the fold is mainly stabilized by the presence of three disulfide bonds. In addition to the overall shape of the molecules, the ratio of solvent-exposed polar and hydrophobic side chains is also very similar among the four defensins investigated. It is significant that beta-defensins do not exhibit a common pattern of charged and hydrophobic residues on the protein surface and that the beta-defensin-specific fold appears to accommodate a wide range of different amino acids at most sequence positions. In addition to the implications for the mode of biological defensin actions, these findings are of particular interest because beta-defensins have been suggested as lead compounds for the development of novel peptide antibiotics for the therapy of infectious diseases.
Figures
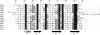


Similar articles
-
Engineering disulfide bonds of the novel human beta-defensins hBD-27 and hBD-28: differences in disulfide formation and biological activity among human beta-defensins.Biopolymers. 2005;80(1):34-49. doi: 10.1002/bip.20193. Biopolymers. 2005. PMID: 15625724
-
Solution structure of bovine neutrophil beta-defensin-12: the peptide fold of the beta-defensins is identical to that of the classical defensins.Biochemistry. 1995 Oct 17;34(41):13663-71. doi: 10.1021/bi00041a048. Biochemistry. 1995. PMID: 7577957
-
Solution structure of spheniscin, a beta-defensin from the penguin stomach.J Biol Chem. 2004 Jul 16;279(29):30433-9. doi: 10.1074/jbc.M401338200. Epub 2004 Apr 30. J Biol Chem. 2004. PMID: 15123713
-
The beta-defensin-fold family of polypeptides.Toxicon. 2004 Nov;44(6):581-8. doi: 10.1016/j.toxicon.2004.07.011. Toxicon. 2004. PMID: 15501283 Review.
-
Human beta-defensins.Cell Mol Life Sci. 2006 Jun;63(11):1294-313. doi: 10.1007/s00018-005-5540-2. Cell Mol Life Sci. 2006. PMID: 16710608 Free PMC article. Review.
Cited by
-
Sequence conservation, phylogenetic relationships, and expression profiles of nondigestive serine proteases and serine protease homologs in Manduca sexta.Insect Biochem Mol Biol. 2015 Jul;62:51-63. doi: 10.1016/j.ibmb.2014.10.006. Epub 2014 Dec 19. Insect Biochem Mol Biol. 2015. PMID: 25530503 Free PMC article.
-
Integrated Transcriptomic and Proteomic Analysis of Red Blood Cells from Rainbow Trout Challenged with VHSV Point Towards Novel Immunomodulant Targets.Vaccines (Basel). 2019 Jul 9;7(3):63. doi: 10.3390/vaccines7030063. Vaccines (Basel). 2019. PMID: 31324030 Free PMC article.
-
Human defensins.J Mol Med (Berl). 2005 Aug;83(8):587-95. doi: 10.1007/s00109-005-0657-1. Epub 2005 Apr 9. J Mol Med (Berl). 2005. PMID: 15821901 Review.
-
Antimicrobial proteins and polypeptides in pulmonary innate defence.Respir Res. 2006 Feb 17;7(1):29. doi: 10.1186/1465-9921-7-29. Respir Res. 2006. PMID: 16503962 Free PMC article. Review.
-
Antimicrobial peptides and the gut microbiome in inflammatory bowel disease.World J Gastroenterol. 2021 Nov 21;27(43):7402-7422. doi: 10.3748/wjg.v27.i43.7402. World J Gastroenterol. 2021. PMID: 34887639 Free PMC article.
References
-
- Baba, M., Imai, T., Nishimura, M., Kakizaki, M., Takagi, S., Hieshima, K., Nomiyama, H., and Yoshie, O. 1997. Identification of CCR6, the specific receptor for a novel lymphocyte-directed CC chemokine LARC. J. Biol. Chem. 272 14893–14898. - PubMed
-
- Barton, G.J. 1993. ALSCRIPT: A tool to format multiple sequence alignments. Protein Eng. 6 37–40. - PubMed
-
- Bensch, K.W., Raida, M., Mägert, H.J., Schulz-Knappe, P., and Forssmann, W.G. 1995. hBD-1: A novel beta-defensin from human plasma. FEBS Lett. 368 331–335. - PubMed
-
- Boman, H.G. 1995. Peptide antibiotics and their role in innate immunity. Annu. Rev. Immunol. 13 61–92. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

