Thermodynamics of interactions of urea and guanidinium salts with protein surface: relationship between solute effects on protein processes and changes in water-accessible surface area
- PMID: 11714916
- PMCID: PMC2374034
- DOI: 10.1110/ps.ps.20801
Thermodynamics of interactions of urea and guanidinium salts with protein surface: relationship between solute effects on protein processes and changes in water-accessible surface area
Abstract
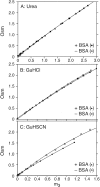
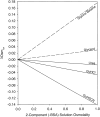
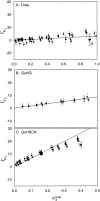
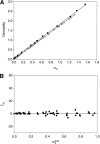
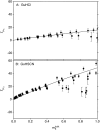
To interpret effects of urea and guanidinium (GuH(+)) salts on processes that involve large changes in protein water-accessible surface area (ASA), and to predict these effects from structural information, a thermodynamic characterization of the interactions of these solutes with different types of protein surface is required. In the present work we quantify the interactions of urea, GuHCl, GuHSCN, and, for comparison, KCl with native bovine serum albumin (BSA) surface, using vapor pressure osmometry (VPO) to obtain preferential interaction coefficients (Gamma(mu3)) as functions of nondenaturing concentrations of these solutes (0-1 molal). From analysis of Gamma(mu3) using the local-bulk domain model, we obtain concentration-independent partition coefficients K(nat)(P) that characterize the accumulation of these solutes near native protein (BSA) surface: K(nat)(P,urea)= 1.10 +/- 0.04, K(nat)(P,SCN(-)) = 2.4 +/- 0.2, K(nat)(P,GuH(+)) = 1.60 +/- 0.08, relative to K(nat)(P,K(+)) identical with 1 and K(nat)(P,Cl(-)) = 1.0 +/- 0.08. The relative magnitudes of K(nat)(P) are consistent with the relative effectiveness of these solutes as perturbants of protein processes. From a comparison of partition coefficients for these solutes and native surface (K(nat)(P)) with those determined by us previously for unfolded protein and alanine-based peptide surface K(unf)(P), we dissect K(P) into contributions from polar peptide backbone and other types of protein surface. For globular protein-urea interactions, we find K(nat)(P,urea) = K(unf)(P,urea). We propose that this equality arises because polar peptide backbone is the same fraction (0.13) of total ASA for both classes of surface. The analysis presented here quantifies and provides a physical basis for understanding Hofmeister effects of salt ions and the effects of uncharged solutes on protein processes in terms of K(P) and the change in protein ASA.
Figures

Similar articles
-
Thermodynamic analysis of interactions between denaturants and protein surface exposed on unfolding: interpretation of urea and guanidinium chloride m-values and their correlation with changes in accessible surface area (ASA) using preferential interaction coefficients and the local-bulk domain model.Proteins. 2000;Suppl 4:72-85. doi: 10.1002/1097-0134(2000)41:4+<72::aid-prot70>3.0.co;2-7. Proteins. 2000. PMID: 11013402
-
Vapor pressure osmometry studies of osmolyte-protein interactions: implications for the action of osmoprotectants in vivo and for the interpretation of "osmotic stress" experiments in vitro.Biochemistry. 2000 Apr 18;39(15):4455-71. doi: 10.1021/bi992887l. Biochemistry. 2000. PMID: 10757995
-
Thermodynamic characterization of interactions of native bovine serum albumin with highly excluded (glycine betaine) and moderately accumulated (urea) solutes by a novel application of vapor pressure osmometry.Biochemistry. 1996 Aug 13;35(32):10506-16. doi: 10.1021/bi960795f. Biochemistry. 1996. PMID: 8756707
-
Water as ligand: preferential binding and exclusion of denaturants in protein unfolding.Biochemistry. 1992 Oct 20;31(41):9857-64. doi: 10.1021/bi00156a001. Biochemistry. 1992. PMID: 1390769 Review. No abstract available.
-
Uncovering the basis for nonideal behavior of biological molecules.Biochemistry. 2004 Nov 16;43(45):14472-84. doi: 10.1021/bi048681o. Biochemistry. 2004. PMID: 15533052 Review.
Cited by
-
Hydration of guanidinium depends on its local environment.Chem Sci. 2015 Jun 1;6(6):3420-3429. doi: 10.1039/c5sc00618j. Epub 2015 Apr 14. Chem Sci. 2015. PMID: 28706704 Free PMC article.
-
Preferential interactions of guanidinum ions with aromatic groups over aliphatic groups.J Am Chem Soc. 2009 Nov 25;131(46):16689-96. doi: 10.1021/ja903478s. J Am Chem Soc. 2009. PMID: 19874022 Free PMC article.
-
Anatomy of energetic changes accompanying urea-induced protein denaturation.Proc Natl Acad Sci U S A. 2007 Sep 25;104(39):15317-22. doi: 10.1073/pnas.0706251104. Epub 2007 Sep 18. Proc Natl Acad Sci U S A. 2007. PMID: 17878304 Free PMC article.
-
Protein stability in mixed solvents: a balance of contact interaction and excluded volume.Biophys J. 2003 Jul;85(1):108-25. doi: 10.1016/S0006-3495(03)74459-2. Biophys J. 2003. PMID: 12829469 Free PMC article.
-
Quantifying Amide-Aromatic Interactions at Molecular and Atomic Levels: Experimentally-determined Enthalpic and Entropic Contributions to Interactions of Amide sp 2 O, N, C and sp 3 C Unified Atoms with Naphthalene sp 2 C Atoms in Water.bioRxiv [Preprint]. 2023 Jul 17:2023.07.12.548600. doi: 10.1101/2023.07.12.548600. bioRxiv. 2023. Update in: Biochemistry. 2023 Oct 3;62(19):2841-2853. doi: 10.1021/acs.biochem.3c00367. PMID: 37503153 Free PMC article. Updated. Preprint.
References
-
- Arakawa, T. and Timasheff, S.N. 1982. Preferential interactions of proteins with salts in concentrated solutions. Biochemistry 21 6545–6552. - PubMed
-
- ———. 1983. Preferential interactions of proteins with solvent components in aqueous amino acid solutions. Arch. Biochem. Biophys. 224 169–177. - PubMed
-
- ———. 1984a. Protein stabilization and destabilization by guanidinium salts. Biochemistry 23 5924–5929. - PubMed
-
- ———. 1984b. The mechanism of action of Na glutamate, lysine HCl, and piperazine-N, N`-bis (2-ethanesulfonic acid) in the stabilization of tubulin and microtubule formation. J. Biol. Chem. 259 4979–4986. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

