The role of strand 1 of the C beta-sheet in the structure and function of alpha(1)-antitrypsin
- PMID: 11714919
- PMCID: PMC2374035
- DOI: 10.1110/ps.ps.24101
The role of strand 1 of the C beta-sheet in the structure and function of alpha(1)-antitrypsin
Abstract
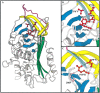
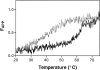
Serpins inhibit cognate serine proteases involved in a number of important processes including blood coagulation and inflammation. Consequently, loss of serpin function or stability results in a number of disease states. Many of the naturally occurring mutations leading to disease are located within strand 1 of the C beta-sheet of the serpin. To ascertain the structural and functional importance of each residue in this strand, which constitutes the so-called distal hinge of the reactive center loop of the serpin, an alanine scanning study was carried out on recombinant alpha(1)-antitrypsin Pittsburgh mutant (P1 = Arg). Mutation of the P10' position had no effect on its inhibitory properties towards thrombin. Mutations to residues P7' and P9' caused these serpins to have an increased tendency to act as substrates rather than inhibitors, while mutations at P6' and P8' positions caused the serpin to behave almost entirely as a substrate. Mutations at the P6' and P8' residues of the C beta-sheet, which are buried in the hydrophobic core in the native structure, caused the serpin to become highly unstable and polymerize much more readily. Thus, P6' and P8' mutants of alpha(1)-antitrypsin had melting temperatures 14 degrees lower than wild-type alpha(1)-antitrypsin. These results indicate the importance of maintaining the anchoring of the distal hinge to both the inhibitory mechanism and stability of serpins, the inhibitory mechanism being particularly sensitive to any perturbations in this region. The results of this study allow more informed analysis of the effects of mutations found at these positions in disease-associated serpin variants.
Figures

Similar articles
-
Modeling of serpin-protease complexes: antithrombin-thrombin, alpha 1-antitrypsin (358Met-->Arg)-thrombin, alpha 1-antitrypsin (358Met-->Arg)-trypsin, and antitrypsin-elastase.Proteins. 1996 Nov;26(3):288-303. doi: 10.1002/(SICI)1097-0134(199611)26:3<288::AID-PROT5>3.0.CO;2-A. Proteins. 1996. PMID: 8953650
-
Preparative induction and characterization of L-antithrombin: a structural homologue of latent plasminogen activator inhibitor-1.Biochemistry. 1997 Oct 21;36(42):13133-42. doi: 10.1021/bi970664u. Biochemistry. 1997. PMID: 9335576
-
Role of the connectivity of secondary structure segments in the folding of alpha(1)-antitrypsin.Biochem Biophys Res Commun. 2001 Sep 28;287(3):636-41. doi: 10.1006/bbrc.2001.5638. Biochem Biophys Res Commun. 2001. PMID: 11563842
-
Wild-type alpha 1-antitrypsin is in the canonical inhibitory conformation.J Mol Biol. 1998 Jan 23;275(3):419-25. doi: 10.1006/jmbi.1997.1458. J Mol Biol. 1998. PMID: 9466920 Review.
-
Mechanisms of serpin dysfunction in disease.Expert Rev Mol Med. 2006 Dec 11;8(31):1-19. doi: 10.1017/S1462399406000184. Expert Rev Mol Med. 2006. PMID: 17156576 Review.
Cited by
-
Local conformational flexibility provides a basis for facile polymer formation in human neuroserpin.Biophys J. 2011 Oct 5;101(7):1758-65. doi: 10.1016/j.bpj.2011.08.037. Biophys J. 2011. PMID: 21961602 Free PMC article.
-
Engineering the serpin α1 -antitrypsin: A diversity of goals and techniques.Protein Sci. 2020 Apr;29(4):856-871. doi: 10.1002/pro.3794. Epub 2019 Dec 9. Protein Sci. 2020. PMID: 31774589 Free PMC article. Review.
-
Phage display of the serpin alpha-1 proteinase inhibitor randomized at consecutive residues in the reactive centre loop and biopanned with or without thrombin.PLoS One. 2014 Jan 10;9(1):e84491. doi: 10.1371/journal.pone.0084491. eCollection 2014. PLoS One. 2014. PMID: 24427287 Free PMC article.
-
alpha(1)-Proteinase inhibitor mutants with specificity for plasma kallikrein and C1s but not C1.Protein Sci. 2002 Sep;11(9):2230-6. doi: 10.1110/ps.0207302. Protein Sci. 2002. PMID: 12192078 Free PMC article.
References
-
- Beauchamp, N.J., Pike, R.N., Daly, M., Butler, L., Makris, M., Dafforn, T.R., Zhou, A., Fitton, H.L., Preston, F.E., Peake, I.R., and Carrell, R.W. 1998. Antithrombins Wibble and Wobble (T85M/K): Archetypal conformational diseases with in vivo latent-transition, thrombosis, and heparin activation. Blood 92 2696–2706. - PubMed
-
- Bock, S.C., Marrinan, J.A., and Radziejewska, E. 1988. Antithrombin III Utah: Proline-407 to leucine mutation in a highly conserved region near the inhibitor reactive site. Biochemistry 27 6171–6178. - PubMed
-
- Bottomley, S.P. and Stone, S.R. 1998. Protein engineering of chimeric Serpins: An investigation into effects of the serpin scaffold and reactive center loop length. Protein Eng. 11 1243–1247. - PubMed
-
- Carrell, R.W. and Gooptu, B. 1998. Conformational changes and disease—Serpins, prions and Alzheimer's. Curr. Opin. Struct. Biol. 8 799–809. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

