Molecular design of Mycoplasma hominis Vaa adhesin
- PMID: 11714926
- PMCID: PMC2374042
- DOI: 10.1110/ps.ps.31901
Molecular design of Mycoplasma hominis Vaa adhesin
Abstract
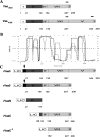
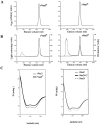
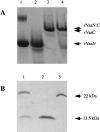
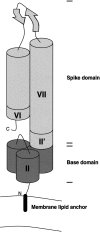
The variable adherence-associated (Vaa) adhesin of the opportunistic human pathogen Mycoplasma hominis is a surface-exposed, membrane-associated protein involved in the attachment of the bacterium to host cells. The molecular masses of recombinant 1 and 2 cassette forms of the protein determined by a light-scattering (LS) method were 23.9 kD and 36.5 kD, respectively, and corresponded to their monomeric forms. Circular dichroism (CD) spectroscopy of the full-length forms indicated that the Vaa protein has an alpha-helical content of approximately 80%. Sequence analysis indicates the presence of coiled-coil domains in both the conserved N-terminal and antigenic variable C-terminal part of the Vaa adhesin. Experimental results obtained with recombinant proteins corresponding to the N- or C-terminal parts of the shortest one-cassette form of the protein were consistent with the hypothesis of two distinct coiled-coil regions. The one-cassette Vaa monomer appears to be an elongated protein with a axial shape ratio of 1:10. Analysis of a two-cassette Vaa type reveals a similar axial shape ratio. The results are interpreted in terms of the topological organization of the Vaa protein indicating the localization of the adherence-mediating structure.
Figures

Similar articles
-
The vaa locus of Mycoplasma hominis contains a divergent genetic islet encoding a putative membrane protein.BMC Microbiol. 2004 Sep 22;4:37. doi: 10.1186/1471-2180-4-37. BMC Microbiol. 2004. PMID: 15385054 Free PMC article.
-
Variability of the Vaa cytoadhesin genes in clinical isolates of Mycoplasma hominis.New Microbiol. 2005 Oct;28(4):373-6. New Microbiol. 2005. PMID: 16386023
-
Localized reversible frameshift mutation in an adhesin gene confers a phase-variable adherence phenotype in mycoplasma.Mol Microbiol. 1997 Sep;25(5):859-69. doi: 10.1111/j.1365-2958.1997.mmi509.x. Mol Microbiol. 1997. PMID: 9364912
-
Molecular dissection of Mycoplasma hominis.APMIS Suppl. 2000;97:1-45. APMIS Suppl. 2000. PMID: 10721331 Review.
-
Molecular biology of Mycoplasma.Wien Klin Wochenschr. 1997 Aug 8;109(14-15):557-61. Wien Klin Wochenschr. 1997. PMID: 9286059 Review.
Cited by
-
[Clinical implications of the genus Mycoplasma].Rev Esp Quimioter. 2021 Jun;34(3):169-184. doi: 10.37201/req/014.2021. Epub 2021 Mar 18. Rev Esp Quimioter. 2021. PMID: 33735544 Free PMC article. Review. Spanish.
-
The vaa locus of Mycoplasma hominis contains a divergent genetic islet encoding a putative membrane protein.BMC Microbiol. 2004 Sep 22;4:37. doi: 10.1186/1471-2180-4-37. BMC Microbiol. 2004. PMID: 15385054 Free PMC article.
-
Unraveling the adaptive strategies of Mycoplasma hominis through proteogenomic profiling of clinical isolates.Front Cell Infect Microbiol. 2024 May 2;14:1398706. doi: 10.3389/fcimb.2024.1398706. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38756231 Free PMC article.
-
A genome-wide investigation of Mycoplasma hominis genes associated with gynecological infections or infertility.Front Microbiol. 2025 Apr 30;16:1561378. doi: 10.3389/fmicb.2025.1561378. eCollection 2025. Front Microbiol. 2025. PMID: 40371111 Free PMC article.
-
Infection strategies of mycoplasmas: Unraveling the panoply of virulence factors.Virulence. 2021 Dec;12(1):788-817. doi: 10.1080/21505594.2021.1889813. Virulence. 2021. PMID: 33704021 Free PMC article. Review.
References
-
- Boesen, T., Emmersen, J., Jensen, L.T., Ladefoged, S.A., Thorsen, P., Birkelund, S., and Christiansen, G. 1998. The Mycoplasma hominis vaa gene displays a mosaic gene structure. Mol. Microbiol. 29 97–110. - PubMed
-
- Burkhard, P., Kammerer, R.A., Steinmetz, M.O., Bourenkov, G.P., and Aebi, U. 2000. The coiled-coil trigger site of the rod domain of cortexillin I unveils a distinct network of interhelical and intrahelical salt bridges. Structure 8 223–230. - PubMed
-
- Cantor, C.R. and Schimmel, P.R. 1980. Biophysical chemistry part II: Techniques for the study of biological structure and function. W.H. Freeman, San Francisco.
-
- Cooper, T.M. and Woody, R.W. 1990. The effect of conformation on the CD of interacting helices: A theoretical study of tropomyosin. Biopolymers 30 657–676. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

