Genomic organization of Tropomodulins 2 and 4 and unusual intergenic and intraexonic splicing of YL-1 and Tropomodulin 4
- PMID: 11716785
- PMCID: PMC59888
- DOI: 10.1186/1471-2164-2-7
Genomic organization of Tropomodulins 2 and 4 and unusual intergenic and intraexonic splicing of YL-1 and Tropomodulin 4
Abstract
Background: The tropomodulins (TMODs) are a family of proteins that cap the pointed ends of actin filaments. Four TMODs have been identified in humans, with orthologs in mice. Mutations in actin or actin-binding proteins have been found to cause several human diseases, ranging from hypertrophic cardiomyopathy to immunodeficiencies such as Wiskott-Aldrich syndrome. We had previously mapped Tropomodulin 2 (TMOD2) to the genomic region containing the gene for amyotrophic lateral sclerosis 5 (ALS5). We determined the genomic structure of Tmod2 in order to better analyze patient DNA for mutations; we also determined the genomic structure of Tropomodulin 4 (TMOD4).
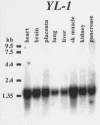
Results: In this study, we determined the genomic structure of TMOD2 and TMOD4 and found the organization of both genes to be similar. Sequence analysis of TMOD2 revealed no mutations or polymorphisms in ALS5 patients or controls. Interestingly, we discovered that another gene, YL-1, intergenically splices into TMOD4. YL-1 encodes six exons, the last of which is 291 bp from a 5' untranslated exon of TMOD4. We used 5' RACE and RT-PCR from TMOD4 to identify several intergenic RACE products. YL-1 was also found to undergo unconventional splicing using non-canonical splice sites within exons (intraexonic splicing) to produce several alternative transcripts.
Conclusions: The genomic structure of TMOD2 and TMOD4 have been delineated. This should facilitate future mutational analysis of these genes. In addition, intergenic splicing at TMOD4/YL-1 was discovered, demonstrating yet another level of complexity of gene organization and regulation.
Figures


References
-
- Sung LA, Fowler VM, Lambert K, Sussman MA, Karr D, Chien S. Molecular cloning and characterization of human fetal liver tropomodulin. A tropomyosin-binding protein. J Biol Chem. 1992;267:2616–2621. - PubMed
-
- Sussman MA, Sakhi S, Barrientos P, Ito M, Kedes L. Tropomodulin in rat cardiac muscle. Localization of protein is independent of messenger RNA distribution during myofibrillar development. Circ Res. 1994;75:221–232. - PubMed
-
- Watakabe A, Kobayashi R, Helfman DM. N-tropomodulin: a novel isoform of tropomodulin identified as the major binding protein to brain tropomyosin. J Cell Sci. 1996;109:2299–2310. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

