DNA microarray analysis of the hyperthermophilic archaeon Pyrococcus furiosus: evidence for anNew type of sulfur-reducing enzyme complex
- PMID: 11717259
- PMCID: PMC95549
- DOI: 10.1128/JB.183.24.7027-7036.2001
DNA microarray analysis of the hyperthermophilic archaeon Pyrococcus furiosus: evidence for anNew type of sulfur-reducing enzyme complex
Abstract
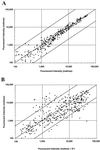
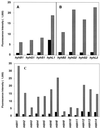
DNA microarrays were constructed by using 271 open reading frame (ORFs) from the genome of the archaeon Pyrococcus furiosus. They were used to investigate the effects of elemental sulfur (S(primary)) on the levels of gene expression in cells grown at 95 degrees C with maltose as the carbon source. The ORFs included those that are proposed to encode proteins mainly involved in the pathways of sugar and peptide catabolism, in the metabolism of metals, and in the biosynthesis of various cofactors, amino acids, and nucleotides. The expression of 21 ORFs decreased by more than fivefold when cells were grown with S(primary) and, of these, 18 encode subunits associated with three different hydrogenase systems. The remaining three ORFs encode homologs of ornithine carbamoyltransferase and HypF, both of which appear to be involved in hydrogenase biosynthesis, as well as a conserved hypothetical protein. The expression of two previously uncharacterized ORFs increased by more than 25-fold when cells were grown with S(primary). Their products, termed SipA and SipB (for sulfur-induced proteins), are proposed to be part of a novel S(primary)-reducing, membrane-associated, iron-sulfur cluster-containing complex. Two other previously uncharacterized ORFs encoding a putative flavoprotein and a second FeS protein were upregulated more than sixfold in S(primary)-grown cells, and these are also thought be involved in S(primary) reduction. Four ORFs that encode homologs of proteins involved in amino acid metabolism were similarly upregulated in S(primary)-grown cells, a finding consistent with the fact that growth on peptides is a S(primary)-dependent process. An ORF encoding a homolog of the eukaryotic rRNA processing protein, fibrillarin, was also upregulated sixfold in the presence of S(primary), although the reason for this is as yet unknown. Of the 20 S(primary)-independent ORFs that are the most highly expressed (at more than 20 times the detection limit), 12 of them represent enzymes purified from P. furiosus, but none of the products of the 34 S(primary)-independent ORFs that are not expressed above the detection limit have been characterized. These results represent the first derived from the application of DNA microarrays to either an archaeon or a hyperthermophile.
Figures

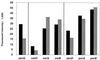
Similar articles
-
The elemental sulfur-responsive protein (SipA) from the hyperthermophilic archaeon Pyrococcus furiosus is regulated by sulfide in an iron-dependent manner.J Bacteriol. 2010 Nov;192(21):5841-3. doi: 10.1128/JB.00660-10. Epub 2010 Aug 27. J Bacteriol. 2010. PMID: 20802041 Free PMC article.
-
Whole-genome DNA microarray analysis of a hyperthermophile and an archaeon: Pyrococcus furiosus grown on carbohydrates or peptides.J Bacteriol. 2003 Jul;185(13):3935-47. doi: 10.1128/JB.185.13.3935-3947.2003. J Bacteriol. 2003. PMID: 12813088 Free PMC article.
-
Insights into the metabolism of elemental sulfur by the hyperthermophilic archaeon Pyrococcus furiosus: characterization of a coenzyme A- dependent NAD(P)H sulfur oxidoreductase.J Bacteriol. 2007 Jun;189(12):4431-41. doi: 10.1128/JB.00031-07. Epub 2007 Apr 20. J Bacteriol. 2007. PMID: 17449625 Free PMC article.
-
The role of TrmB and TrmB-like transcriptional regulators for sugar transport and metabolism in the hyperthermophilic archaeon Pyrococcus furiosus.Arch Microbiol. 2008 Sep;190(3):247-56. doi: 10.1007/s00203-008-0378-2. Epub 2008 May 11. Arch Microbiol. 2008. PMID: 18470695 Review.
-
Biochemical diversity among sulfur-dependent, hyperthermophilic microorganisms.FEMS Microbiol Rev. 1994 Oct;15(2-3):261-77. doi: 10.1111/j.1574-6976.1994.tb00139.x. FEMS Microbiol Rev. 1994. PMID: 7946471 Review.
Cited by
-
Molecular and biochemical characterization of a distinct type of fructose-1,6-bisphosphatase from Pyrococcus furiosus.J Bacteriol. 2002 Jun;184(12):3401-5. doi: 10.1128/JB.184.12.3401-3405.2002. J Bacteriol. 2002. PMID: 12029059 Free PMC article.
-
A simple energy-conserving system: proton reduction coupled to proton translocation.Proc Natl Acad Sci U S A. 2003 Jun 24;100(13):7545-50. doi: 10.1073/pnas.1331436100. Epub 2003 Jun 5. Proc Natl Acad Sci U S A. 2003. PMID: 12792025 Free PMC article.
-
The elemental sulfur-responsive protein (SipA) from the hyperthermophilic archaeon Pyrococcus furiosus is regulated by sulfide in an iron-dependent manner.J Bacteriol. 2010 Nov;192(21):5841-3. doi: 10.1128/JB.00660-10. Epub 2010 Aug 27. J Bacteriol. 2010. PMID: 20802041 Free PMC article.
-
Production and characterization of a thermostable alcohol dehydrogenase that belongs to the aldo-keto reductase superfamily.Appl Environ Microbiol. 2006 Jan;72(1):233-8. doi: 10.1128/AEM.72.1.233-238.2006. Appl Environ Microbiol. 2006. PMID: 16391048 Free PMC article.
-
SurR: a transcriptional activator and repressor controlling hydrogen and elemental sulphur metabolism in Pyrococcus furiosus.Mol Microbiol. 2009 Jan;71(2):332-49. doi: 10.1111/j.1365-2958.2008.06525.x. Epub 2008 Nov 10. Mol Microbiol. 2009. PMID: 19017274 Free PMC article.
References
-
- Adams M W W, Holden J F, Menon A L, Schut G J, Grunden A M, Hou C, Hutchins A M, Jenney F E, Jr, Kim C, Ma K, Pan G, Roy R, Sapra R, Story S V, Verhagen M F. Key role for sulfur in peptide metabolism and in regulation of three hydrogenases in the hyperthermophilic archaeon Pyrococcus furiosus. J Bacteriol. 2001;183:716–724. - PMC - PubMed
-
- Arfin S M, Long A D, Ito E T, Tolleri L, Riehle M M, Paegle E S, Hatfield G W. Global gene expression profiling in Escherichia coli K12. The effects of integration host factor. J Biol Chem. 2000;275:29672–29684. - PubMed
-
- Barbier G, Godfroy A, Meunier J R, Querellou J, Cambon M A, Lesongeur F, Grimont P A, Raguenes G. Pyrococcus glycovorans sp. nov., a hyperthermophilic archaeon isolated from the East Pacific Rise. Int J Syst Bacteriol. 1999;49(Pt. 4):1829–1837. - PubMed
-
- Blamey J, Chiong M, Lopez C, Smith E. Optimization of the growth conditions of the extremely thermophilic microorganisms Thermococcus celer and Pyrococcus woesei. J Microbiol Methods. 1999;38:169–175. - PubMed
-
- Blamey J M, Adams M W W. Characterization of an ancestral type of pyruvate ferredoxin oxidoreductase from the hyperthermophilic bacterium Thermotoga maritima. Biochemistry. 1994;33:1000–1007. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

