Use of transposon Tn5367 mutagenesis and a nitroimidazopyran-based selection system to demonstrate a requirement for fbiA and fbiB in coenzyme F(420) biosynthesis by Mycobacterium bovis BCG
- PMID: 11717263
- PMCID: PMC95553
- DOI: 10.1128/JB.183.24.7058-7066.2001
Use of transposon Tn5367 mutagenesis and a nitroimidazopyran-based selection system to demonstrate a requirement for fbiA and fbiB in coenzyme F(420) biosynthesis by Mycobacterium bovis BCG
Abstract
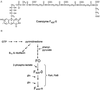
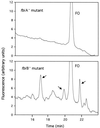
Three transposon Tn5367 mutagenesis vectors (phAE94, pPR28, and pPR29) were used to create a collection of insertion mutants of Mycobacterium bovis strain BCG. A strategy to select for transposon-generated mutants that cannot make coenzyme F(420) was developed using the nitroimidazopyran-based antituberculosis drug PA-824. One-third of 134 PA-824-resistant mutants were defective in F(420) accumulation. Two mutants that could not make F(420)-5,6 but which made the biosynthesis intermediate FO were examined more closely. These mutants contained transposons inserted in two adjacent homologues of Mycobacterium tuberculosis genes, which we have named fbiA and fbiB for F(420) biosynthesis. Homologues of fbiA were found in all seven microorganisms that have been fully sequenced and annotated and that are known to make F(420). fbiB homologues were found in all but one such organism. Complementation of the fbiA mutant with fbiAB and complementation of the fbiB mutant with fbiB both restored the F(420)-5,6 phenotype. Complementation of the fbiA mutant with fbiA or fbiB alone did not restore the F(420)-5,6 phenotype, but the fbiA mutant complemented with fbiA produced F(420)-2,3,4 at levels similar to F(420)-5,6 made by the wild-type strain, but produced much less F(420)-5. These data demonstrate that both genes are essential for normal F(420)-5,6 production and suggest that the fbiA mutation has a partial polar effect on fbiB. Reverse transcription-PCR data demonstrated that fbiA and fbiB constitute an operon. However, very low levels of fbiB mRNA are produced by the fbiA mutant, suggesting that a low-level alternative start site is located upstream of fbiB. The specific reactions catalyzed by FbiA and FbiB are unknown, but both function between FO and F(420)-5,6, since FO is made by both mutants.
Figures


References
-
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Bair T B. Methods to identify genes involved in F420 biosynthesis by Mycobacterium. Ph.D. thesis. Iowa City: University of Iowa; 2001.
-
- Bair T B, Isabelle D W, Daniels L. Structures of coenzyme F420 in Mycobacterium species. Arch Microbiol. 2001;176:37–43. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

