Characterization of flagellum gene families of methanogenic archaea and localization of novel flagellum accessory proteins
- PMID: 11717274
- PMCID: PMC95564
- DOI: 10.1128/JB.183.24.7154-7164.2001
Characterization of flagellum gene families of methanogenic archaea and localization of novel flagellum accessory proteins
Abstract
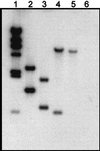
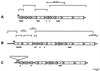
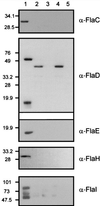
Archaeal flagella are unique motility structures, and the absence of bacterial structural motility genes in the complete genome sequences of flagellated archaeal species suggests that archaeal flagellar biogenesis is likely mediated by novel components. In this study, a conserved flagellar gene family from each of Methanococcus voltae, Methanococcus maripaludis, Methanococcus thermolithotrophicus, and Methanococcus jannaschii has been characterized. These species possess multiple flagellin genes followed immediately by eight known and supposed flagellar accessory genes, flaCDEFGHIJ. Sequence analyses identified a conserved Walker box A motif in the putative nucleotide binding proteins FlaH and FlaI that may be involved in energy production for flagellin secretion or assembly. Northern blotting studies demonstrated that all the species have abundant polycistronic mRNAs corresponding to some of the structural flagellin genes, and in some cases several flagellar accessory genes were shown to be cotranscribed with the flagellin genes. Cloned flagellar accessory genes of M. voltae were successfully overexpressed as His-tagged proteins in Escherichia coli. These recombinant flagellar accessory proteins were affinity purified and used as antigens to raise polyclonal antibodies for localization studies. Immunoblotting of fractionated M. voltae cells demonstrated that FlaC, FlaD, FlaE, FlaH, and FlaI are all present in the cell as membrane-associated proteins but are not major components of isolated flagellar filaments. Interestingly, flaD was found to encode two proteins, each translated from a separate ribosome binding site. These protein expression data indicate for the first time that the putative flagellar accessory genes of M. voltae, and likely those of other archaeal species, do encode proteins that can be detected in the cell.
Figures



Similar articles
-
Systematic deletion analyses of the fla genes in the flagella operon identify several genes essential for proper assembly and function of flagella in the archaeon, Methanococcus maripaludis.Mol Microbiol. 2007 Nov;66(3):596-609. doi: 10.1111/j.1365-2958.2007.05913.x. Epub 2007 Sep 20. Mol Microbiol. 2007. PMID: 17887963
-
Insertional inactivation of the flaH gene in the archaeon Methanococcus voltae results in non-flagellated cells.Mol Genet Genomics. 2001 Jun;265(4):596-603. doi: 10.1007/s004380100451. Mol Genet Genomics. 2001. PMID: 11459179
-
Mutants in flaI and flaJ of the archaeon Methanococcus voltae are deficient in flagellum assembly.Mol Microbiol. 2002 Nov;46(3):879-87. doi: 10.1046/j.1365-2958.2002.03220.x. Mol Microbiol. 2002. PMID: 12410843
-
Archaeal flagella, bacterial flagella and type IV pili: a comparison of genes and posttranslational modifications.J Mol Microbiol Biotechnol. 2006;11(3-5):167-91. doi: 10.1159/000094053. J Mol Microbiol Biotechnol. 2006. PMID: 16983194 Review.
-
Recent advances in the structure and assembly of the archaeal flagellum.J Mol Microbiol Biotechnol. 2004;7(1-2):41-51. doi: 10.1159/000077868. J Mol Microbiol Biotechnol. 2004. PMID: 15170402 Review.
Cited by
-
Insights into FlaI functions in archaeal motor assembly and motility from structures, conformations, and genetics.Mol Cell. 2013 Mar 28;49(6):1069-82. doi: 10.1016/j.molcel.2013.01.014. Epub 2013 Feb 14. Mol Cell. 2013. PMID: 23416110 Free PMC article.
-
Cloning and characterization of archaeal type I signal peptidase from Methanococcus voltae.J Bacteriol. 2003 Oct;185(20):5936-42. doi: 10.1128/JB.185.20.5936-5942.2003. J Bacteriol. 2003. PMID: 14526003 Free PMC article.
-
A Methanogenic Consortium Was Active and Exhibited Long-Term Survival in an Extremely Acidified Thermophilic Bioreactor.Front Microbiol. 2019 Nov 26;10:2757. doi: 10.3389/fmicb.2019.02757. eCollection 2019. Front Microbiol. 2019. PMID: 32038509 Free PMC article.
-
Haloferax volcanii cells lacking the flagellin FlgA2 are hypermotile.Microbiology (Reading). 2013 Nov;159(Pt 11):2249-2258. doi: 10.1099/mic.0.069617-0. Epub 2013 Aug 29. Microbiology (Reading). 2013. PMID: 23989184 Free PMC article.
-
Shaping the archaeal cell envelope.Archaea. 2010 Jul 7;2010:608243. doi: 10.1155/2010/608243. Archaea. 2010. PMID: 20671907 Free PMC article. Review.
References
-
- Auer J, Spicker G, Böck A. Organization and structure of the Methanococcus transcriptional unit homologous to the Escherichia coli “spectinomycin operon”: implications for the evolutionary relationship of 70S and 80S ribosomes. J Mol Biol. 1989;209:21–36. - PubMed
-
- Bayley D P, Florian V, Klein A, Jarrell K F. Flagellin genes of Methanococcus vannielii: amplification by the polymerase chain reaction, demonstration of signal peptides and identification of major components of the flagellar filament. Mol Gen Genet. 1998;258:639–645. - PubMed
-
- Bayley D P, Jarrell K F. Further evidence to suggest that archaeal flagella are related to bacterial type IV pili. J Mol Evol. 1998;46:370–373. - PubMed
-
- Bayley D P, Kalmokoff M L, Jarrell K F. Effect of bacitracin on flagellar assembly and presumed glycosylation of the flagellins of Methanococcus deltae. Arch Microbiol. 1993;160:179–185.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources

