Conversion of pipecolic acid into lysine in Penicillium chrysogenum requires pipecolate oxidase and saccharopine reductase: characterization of the lys7 gene encoding saccharopine reductase
- PMID: 11717275
- PMCID: PMC95565
- DOI: 10.1128/JB.183.24.7165-7172.2001
Conversion of pipecolic acid into lysine in Penicillium chrysogenum requires pipecolate oxidase and saccharopine reductase: characterization of the lys7 gene encoding saccharopine reductase
Abstract
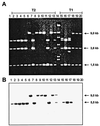
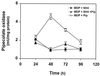
Pipecolic acid is a component of several secondary metabolites in plants and fungi. This compound is useful as a precursor of nonribosomal peptides with novel pharmacological activities. In Penicillium chrysogenum pipecolic acid is converted into lysine and complements the lysine requirement of three different lysine auxotrophs with mutations in the lys1, lys2, or lys3 genes allowing a slow growth of these auxotrophs. We have isolated two P. chrysogenum mutants, named 7.2 and 10.25, that are unable to convert pipecolic acid into lysine. These mutants lacked, respectively, the pipecolate oxidase that converts pipecolic acid into piperideine-6-carboxylic acid and the saccharopine reductase that catalyzes the transformation of piperideine-6-carboxylic acid into saccharopine. The 10.25 mutant was unable to grow in Czapek medium supplemented with alpha-aminoadipic acid. A DNA fragment complementing the 10.25 mutation has been cloned; sequence analysis of the cloned gene (named lys7) revealed that it encoded a protein with high similarity to the saccharopine reductase from Neurospora crassa, Magnaporthe grisea, Saccharomyces cerevisiae, and Schizosaccharomyces pombe. Complementation of the 10.25 mutant with the cloned gene restored saccharopine reductase activity, confirming that lys7 encodes a functional saccharopine reductase. Our data suggest that in P. chrysogenum the conversion of pipecolic acid into lysine proceeds through the transformation of pipecolic acid into piperideine-6-carboxylic acid, saccharopine, and lysine by the consecutive action of pipecolate oxidase, saccharopine reductase, and saccharopine dehydrogenase.
Figures




Similar articles
-
Inactivation of the lys7 gene, encoding saccharopine reductase in Penicillium chrysogenum, leads to accumulation of the secondary metabolite precursors piperideine-6-carboxylic acid and pipecolic acid from alpha-aminoadipic acid.Appl Environ Microbiol. 2004 Feb;70(2):1031-9. doi: 10.1128/AEM.70.2.1031-1039.2004. Appl Environ Microbiol. 2004. PMID: 14766586 Free PMC article.
-
Transcriptional and bioinformatic analysis of the 56.8 kb DNA region amplified in tandem repeats containing the penicillin gene cluster in Penicillium chrysogenum.Fungal Genet Biol. 2006 Sep;43(9):618-29. doi: 10.1016/j.fgb.2006.03.001. Epub 2006 May 18. Fungal Genet Biol. 2006. PMID: 16713314
-
Role of pipecolic acid in the biosynthesis of lysine in Rhodotorula glutinis.J Bacteriol. 1979 May;138(2):410-7. doi: 10.1128/jb.138.2.410-417.1979. J Bacteriol. 1979. PMID: 571433 Free PMC article.
-
alpha-Aminoadipate pathway for the biosynthesis of lysine in lower eukaryotes.Crit Rev Microbiol. 1985;12(2):131-51. doi: 10.3109/10408418509104427. Crit Rev Microbiol. 1985. PMID: 3928261 Review.
-
The alpha-aminoadipate pathway for lysine biosynthesis in fungi.Cell Biochem Biophys. 2006;46(1):43-64. doi: 10.1385/CBB:46:1:43. Cell Biochem Biophys. 2006. PMID: 16943623 Review.
Cited by
-
Penicillium chrysogenum, a Vintage Model with a Cutting-Edge Profile in Biotechnology.Microorganisms. 2022 Mar 6;10(3):573. doi: 10.3390/microorganisms10030573. Microorganisms. 2022. PMID: 35336148 Free PMC article. Review.
-
Regulation and compartmentalization of β-lactam biosynthesis.Microb Biotechnol. 2010 May;3(3):285-99. doi: 10.1111/j.1751-7915.2009.00123.x. Epub 2009 May 31. Microb Biotechnol. 2010. PMID: 21255328 Free PMC article. Review.
-
Isolation of autochthonous non-white rot fungi with potential for enzymatic upgrading of Venezuelan extra-heavy crude oil.Biocatal Biotransformation. 2007 Mar;25(2-4):341-349. doi: 10.1080/10242420701379908. Biocatal Biotransformation. 2007. PMID: 18833334 Free PMC article.
-
Functional analysis through site-directed mutations and phylogeny of the Candida albicans LYS1-encoded saccharopine dehydrogenase.Mol Genet Genomics. 2006 Jan;275(1):74-80. doi: 10.1007/s00438-005-0062-z. Epub 2005 Nov 15. Mol Genet Genomics. 2006. PMID: 16292576
-
Unbiased characterization of genotype-dependent metabolic regulations by metabolomic approach in Arabidopsis thaliana.BMC Syst Biol. 2007 Nov 21;1:53. doi: 10.1186/1752-0509-1-53. BMC Syst Biol. 2007. PMID: 18028551 Free PMC article.
References
-
- Aharonowitz Y, Cohen G, Martín J F. Penicillin and cephalosporin biosynthetic genes: structure, organization, regulation and evolution. Annu Rev Microbiol. 1992;46:461–495. - PubMed
-
- Aspen A J, Meister A. Conversion of α-aminoadipic acid to L-pipecolic acid by Aspergillus nidulans. Biochemistry. 1962;1:606–611. - PubMed
-
- Bañuelos O, Casqueiro J, Fierro F, Hijarrubia M J, Gutiérrez S, Martín J F. Characterization and lysine control of the expression of the lys1 gene encoding homocitrate synthase. Gene. 1999;226:51–59. - PubMed
-
- Bañuelos O, Casqueiro J, Gutiérrez S, Martín J F. Overexpression of the lys1 gene in Penicillium chrysogenum: homocitrate synthase levels, α-aminoadipic acid pool and penicillin production. Appl Microbiol Biotechnol. 2000;54:69–77. - PubMed
-
- Bañuelos O, Casqueiro J, Gutiérrez S, Riaño J, Martín J F. The specific transport system for lysine is fully inhibited by ammonium in Penicillium chrysogenum: an ammonium-insensitive system allows uptake in carbon-starved cells. Antonie Leeuwenhoek. 2000;77:91–100. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

