Correlation between Bacillus subtilis scoC phenotype and gene expression determined using microarrays for transcriptome analysis
- PMID: 11717292
- PMCID: PMC95582
- DOI: 10.1128/JB.183.24.7329-7340.2001
Correlation between Bacillus subtilis scoC phenotype and gene expression determined using microarrays for transcriptome analysis
Abstract
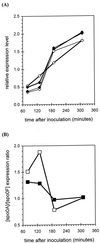
The availability of the complete sequence of the Bacillus subtilis chromosome (F. Kunst et al., Nature 390:249-256, 1997) makes possible the construction of genome-wide DNA arrays and the study of this organism on a global scale. Because we have a long-standing interest in the effects of scoC on late-stage developmental phenomena as they relate to aprE expression, we studied the genome-wide effects of a scoC null mutant with the goal of furthering the understanding of the role of scoC in growth and developmental processes. In the present work we compared the expression patterns of isogenic B. subtilis strains, one of which carries a null mutation in the scoC locus (scoC4). The results obtained indicate that scoC regulates, either directly or indirectly, the expression of at least 560 genes in the B. subtilis genome. ScoC appeared to repress as well as activate gene expression. Changes in expression were observed in genes encoding transport and binding proteins, those involved in amino acid, carbohydrate, and nucleotide and/or nucleoside metabolism, and those associated with motility, sporulation, and adaptation to atypical conditions. Changes in gene expression were also observed for transcriptional regulators, along with sigma factors, regulatory phosphatases and kinases, and members of sensor regulator systems. In this report, we discuss some of the phenotypes associated with the scoC mutant in light of the transcriptome changes observed.
Figures

References
-
- Asai K, Takamatsu H, Iwano M, Kodama T, Watabe K, Ogasawara N. The Bacillus subtilis yabQ gene is essential for formation of the spore cortex. Microbiology. 2001;147:919–927. - PubMed
-
- Dod B, Balassa G, Raulet E, Jeannoda V. Spore control (Sco) mutations in Bacillus subtilis. II. Sporulation and the production of extracellular proteases and amylases by Sco mutants. Mol Gen Genet. 1978;163:45–56.
-
- Dowds B C, Hoch J A. Regulation of the oxidative stress response by the hpr gene in Bacillus subtilis. J Gen Microbiol. 1991;137:1121–1125. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

