Translation of a nonpolyadenylated viral RNA is enhanced by binding of viral coat protein or polyadenylation of the RNA
- PMID: 11717411
- PMCID: PMC64674
- DOI: 10.1073/pnas.251542798
Translation of a nonpolyadenylated viral RNA is enhanced by binding of viral coat protein or polyadenylation of the RNA
Abstract
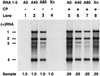
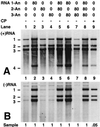
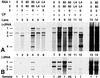
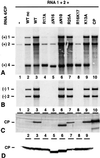
On entering a host cell, positive-strand RNA virus genomes have to serve as messenger for the translation of viral proteins. Efficient translation of cellular messengers requires interactions between initiation factors bound to the 5'-cap structure and the poly(A) binding protein bound to the 3'-poly(A) tail. Initiation of infection with the tripartite RNA genomes of alfalfa mosaic virus (AMV) and viruses from the genus Ilarvirus requires binding of a few molecules of coat protein (CP) to the 3' end of the nonpolyadenylated viral RNAs. Moreover, infection with the genomic RNAs can be initiated by addition of the subgenomic messenger for CP, RNA 4. We report here that extension of the AMV RNAs with a poly(A) tail of 40 to 80 A-residues permitted initiation of infection independently of CP or RNA 4 in the inoculum. Specifically, polyadenylation of RNA 1 relieved an apparent bottleneck in the translation of the viral RNAs. Translation of RNA 4 in plant protoplasts was autocatalytically stimulated by its encoded CP. Mutations that interfered with CP binding to the 3' end of viral RNAs reduced translation of RNA 4 to undetectable levels. Possibly, CP of AMV and ilarviruses stimulates translation of viral RNAs by acting as a functional analogue of poly(A) binding protein or other cellular proteins.
Figures
Similar articles
-
Genetic dissection of the multiple functions of alfalfa mosaic virus coat protein in viral RNA replication, encapsidation, and movement.Virology. 2000 Mar 1;268(1):29-40. doi: 10.1006/viro.1999.0170. Virology. 2000. PMID: 10683324
-
Cis-acting functions of alfalfa mosaic virus proteins involved in replication and encapsidation of viral RNA.Virology. 1999 Feb 15;254(2):324-33. doi: 10.1006/viro.1998.9568. Virology. 1999. PMID: 9986798
-
Replication of alfamo- and ilarviruses: role of the coat protein.Annu Rev Phytopathol. 2005;43:39-62. doi: 10.1146/annurev.phyto.43.101804.120505. Annu Rev Phytopathol. 2005. PMID: 16078876 Review.
-
Coat protein enhances translational efficiency of Alfalfa mosaic virus RNAs and interacts with the eIF4G component of initiation factor eIF4F.J Gen Virol. 2005 Jun;86(Pt 6):1841-1849. doi: 10.1099/vir.0.80796-0. J Gen Virol. 2005. PMID: 15914864
-
Structural and mechanistic insights into hepatitis C viral translation initiation.Nat Rev Microbiol. 2007 Jan;5(1):29-38. doi: 10.1038/nrmicro1558. Epub 2006 Nov 27. Nat Rev Microbiol. 2007. PMID: 17128284 Review.
Cited by
-
Virus versus host cell translation love and hate stories.Adv Virus Res. 2009;73:99-170. doi: 10.1016/S0065-3527(09)73003-9. Adv Virus Res. 2009. PMID: 19695382 Free PMC article. Review.
-
Non-encapsidation activities of the capsid proteins of positive-strand RNA viruses.Virology. 2013 Nov;446(1-2):123-32. doi: 10.1016/j.virol.2013.07.023. Epub 2013 Aug 27. Virology. 2013. PMID: 24074574 Free PMC article. Review.
-
cis-acting elements required for efficient packaging of brome mosaic virus RNA3 in barley protoplasts.J Virol. 2003 Sep;77(18):9979-86. doi: 10.1128/jvi.77.18.9979-9986.2003. J Virol. 2003. PMID: 12941908 Free PMC article.
-
Long-range RNA-RNA interactions circularize the dengue virus genome.J Virol. 2005 Jun;79(11):6631-43. doi: 10.1128/JVI.79.11.6631-6643.2005. J Virol. 2005. PMID: 15890901 Free PMC article.
-
Different Degrees of 5'-to-3' DAR Interactions Modulate Zika Virus Genome Cyclization and Host-Specific Replication.J Virol. 2020 Feb 14;94(5):e01602-19. doi: 10.1128/JVI.01602-19. Print 2020 Feb 14. J Virol. 2020. PMID: 31826997 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

