Conservation of methylation reprogramming in mammalian development: aberrant reprogramming in cloned embryos
- PMID: 11717434
- PMCID: PMC61110
- DOI: 10.1073/pnas.241522698
Conservation of methylation reprogramming in mammalian development: aberrant reprogramming in cloned embryos
Abstract
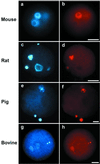
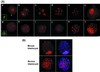
Mouse embryos undergo genome-wide methylation reprogramming by demethylation in early preimplantation development, followed by remethylation thereafter. Here we show that genome-wide reprogramming is conserved in several mammalian species and ask whether it also occurs in embryos cloned with the use of highly methylated somatic donor nuclei. Normal bovine, rat, and pig zygotes showed a demethylated paternal genome, suggesting active demethylation. In bovine embryos methylation was further reduced during cleavage up to the eight-cell stage, and this reduction in methylation was followed by de novo methylation by the 16-cell stage. In cloned one-cell embryos there was a reduction in methylation consistent with active demethylation, but no further demethylation occurred subsequently. Instead, de novo methylation and nuclear reorganization of methylation patterns resembling those of differentiated cells occurred precociously in many cloned embryos. Cloned, but not normal, morulae had highly methylated nuclei in all blastomeres that resembled those of the fibroblast donor cells. Our study shows that epigenetic reprogramming occurs aberrantly in most cloned embryos; incomplete reprogramming may contribute to the low efficiency of cloning.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

